Difference between revisions of "2010 Summer Project Week Cortical Thickness Analysis"
From NAMIC Wiki
| (9 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
Image:PW-MIT2010.png|[[2010_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2010.png|[[2010_Summer_Project_Week#Projects|Projects List]] | ||
Image:Arctic_Logo.png|[http://www.nitrc.org/projects/arctic ARCTIC: Automatic Regional Cortical ThICkness] | Image:Arctic_Logo.png|[http://www.nitrc.org/projects/arctic ARCTIC: Automatic Regional Cortical ThICkness] | ||
| − | Image: | + | Image:GAMBITLogo.png|[http://www.nitrc.org/projects/gambit GAMBIT: Group-wise Automatic Mesh-Based analysis of cortIcal Thickness] |
| − | |||
| − | |||
Image:MeshBasedCortThick_Particles.jpg|Particles on inflated genus-zero cortical surface | Image:MeshBasedCortThick_Particles.jpg|Particles on inflated genus-zero cortical surface | ||
</gallery> | </gallery> | ||
| + | {| | ||
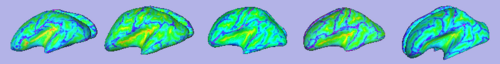
| + | | [[Image:GAMBIT_QC_CorticalThickness.png|thumb|500px|Cortical thickness overlayed on inflated cortical surfaces]] | ||
| + | | [[Image:GAMBIT_QC_SulcalDepth.png|thumb|500px|Sulcal depth overlayed on inflated cortical surfaces]] | ||
| + | |} | ||
| + | ==Key Investigators== | ||
| + | * UNC: Clement Vachet, Heather Cody Hazlett, Martin Styner | ||
| − | == | + | <div style="margin: 20px;"> |
| − | + | <div style="width: 27%; float: left; padding-right: 3%;"> | |
| − | |||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | We would like to create end-to-end applications within Slicer3 allowing voxel-based and mesh-based group analyses of cortical thickness. | |
| + | Such a workflow applied to the young brain (2-4 years old) is our goal in order to start a longitudinal study of early brain development in [[DBP2:UNC|autism (UNC DBP)]]. | ||
| + | |||
| + | See our [[DBP2:UNC:Cortical_Thickness_Roadmap| Roadmap]] for more details. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | |||
| − | |||
| − | |||
| − | + | Our approach is described in the following wiki pages: | |
| − | + | *[[DBP2:UNC:Regional_Cortical_Thickness_Pipeline|Voxel-based regional cortical thickness analysis]] | |
| + | *[[DBP2:UNC:Local_Cortical_Thickness_Pipeline|Mesh-based local cortical thickness analysis]] | ||
| − | ** | + | Two 3D Slicer high-level modules have been developped in that regard: |
| − | + | *[http://www.nitrc.org/projects/arctic ARCTIC: Automatic Regional Cortical ThICkness] | |
| + | *[http://www.nitrc.org/projects/gambit GAMBIT: Group-wise Automatic Mesh-Based analysis of cortIcal Thickness ] | ||
| + | |||
| + | Our plan for the project week is to: | ||
| + | * Follow-up on ARCTIC: Make it directly available as a 3D Slicer extension on MAC (compilation issues apparently) | ||
| + | * Follow-up on GAMBIT: further developmenent | ||
| + | ** Prodiving new 3D Slicer MRML scenes for improved quality control | ||
| + | ** Meet with Manasi Datar and Josh Cates (Utah U.) regarding new features on ShapeWorks (future use of template/sample dataset with fixed particles...) | ||
</div> | </div> | ||
| Line 36: | Line 54: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | * ARCTIC now available as a Slicer3 extension on mac | ||
| + | * GAMBIT: | ||
| + | ** MRML scenes creation with snapshots for improved QC (see pics above) | ||
| + | ** Source code development: other options... | ||
| + | * Particle correspondence: met with Manasi and Beatriz to include new features | ||
| + | * Attended QA breakout session: more tests with code coverage to be added soon on both applications! | ||
| + | * Attended DTI breakout session to identify steps needed to get a consistent package | ||
| Line 46: | Line 71: | ||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
| − | This work will be delivered as | + | This work will be delivered to the NA-MIC Kit as a: |
| + | |||
| + | #ITK Module | ||
| + | #Slicer Module | ||
| + | ##Built-in | ||
| + | ##Extension -- commandline - YES | ||
| + | ##Extension -- loadable | ||
| + | #Other (Please specify) | ||
==References== | ==References== | ||
| − | ** M. Styner, R. | + | *I. Oguz, M. Niethammer, J. Cates, R. Whitaker, T. Fletcher, C. Vachet, and M. Styner, Cortical Correspondence with Probabilistic Fiber Connectivity, Information Processing in Medical Imaging, IPMI 2009, LNCS, in print. |
| + | *H.C. Hazlett, C. Vachet, C. Mathieu, M. Styner, J. Piven, Use of the Slicer3 Toolkit to Produce Regional Cortical Thickness Measurement of Pediatric MRI Data, presented at the 8th Annual International Meeting for Autism Research (IMFAR) Chicago, IL 2009. | ||
| + | *C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2010 abstract | ||
| + | *C. Mathieu, C. Vachet, H.C. Hazlett, G. Geric, J. Piven, and M. Styner, ARCTIC – Automatic Regional Cortical ThICkness Tool, UNC Radiology Research Day 2009 abstract | ||
| + | *J Cates, P T Fletcher, M Styner, M Shenton, R Whitaker. Shape Modeling and Analysis with Entropy-Based Particle Systems. Information Processing in Medical Imaging IPMI 2007, LNCS 4584, pp. 333-345, 2007 | ||
</div> | </div> | ||
Latest revision as of 20:42, 13 December 2010
Home < 2010 Summer Project Week Cortical Thickness AnalysisKey Investigators
- UNC: Clement Vachet, Heather Cody Hazlett, Martin Styner
Objective
We would like to create end-to-end applications within Slicer3 allowing voxel-based and mesh-based group analyses of cortical thickness.
Such a workflow applied to the young brain (2-4 years old) is our goal in order to start a longitudinal study of early brain development in autism (UNC DBP).
See our Roadmap for more details.
Approach, Plan
Our approach is described in the following wiki pages:
Two 3D Slicer high-level modules have been developped in that regard:
- ARCTIC: Automatic Regional Cortical ThICkness
- GAMBIT: Group-wise Automatic Mesh-Based analysis of cortIcal Thickness
Our plan for the project week is to:
- Follow-up on ARCTIC: Make it directly available as a 3D Slicer extension on MAC (compilation issues apparently)
- Follow-up on GAMBIT: further developmenent
- Prodiving new 3D Slicer MRML scenes for improved quality control
- Meet with Manasi Datar and Josh Cates (Utah U.) regarding new features on ShapeWorks (future use of template/sample dataset with fixed particles...)
Progress
- ARCTIC now available as a Slicer3 extension on mac
- GAMBIT:
- MRML scenes creation with snapshots for improved QC (see pics above)
- Source code development: other options...
- Particle correspondence: met with Manasi and Beatriz to include new features
- Attended QA breakout session: more tests with code coverage to be added soon on both applications!
- Attended DTI breakout session to identify steps needed to get a consistent package
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a:
- ITK Module
- Slicer Module
- Built-in
- Extension -- commandline - YES
- Extension -- loadable
- Other (Please specify)
References
- I. Oguz, M. Niethammer, J. Cates, R. Whitaker, T. Fletcher, C. Vachet, and M. Styner, Cortical Correspondence with Probabilistic Fiber Connectivity, Information Processing in Medical Imaging, IPMI 2009, LNCS, in print.
- H.C. Hazlett, C. Vachet, C. Mathieu, M. Styner, J. Piven, Use of the Slicer3 Toolkit to Produce Regional Cortical Thickness Measurement of Pediatric MRI Data, presented at the 8th Annual International Meeting for Autism Research (IMFAR) Chicago, IL 2009.
- C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2010 abstract
- C. Mathieu, C. Vachet, H.C. Hazlett, G. Geric, J. Piven, and M. Styner, ARCTIC – Automatic Regional Cortical ThICkness Tool, UNC Radiology Research Day 2009 abstract
- J Cates, P T Fletcher, M Styner, M Shenton, R Whitaker. Shape Modeling and Analysis with Entropy-Based Particle Systems. Information Processing in Medical Imaging IPMI 2007, LNCS 4584, pp. 333-345, 2007