Difference between revisions of "2010 Summer Project Week Fiducial Deformable Registration"
| (4 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2010.png|[[2010_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2010.png|[[2010_Summer_Project_Week#Projects|Projects List]] | ||
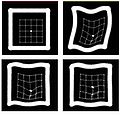
| − | Image: | + | Image:fig-registr-wiki.jpg|Top: Reference and test synthetic images. Bottom: Results for registration without landmark information and using one landmark. |
| − | Image: | + | |
| + | Image:spring.png|Registration error vs number of landmarks. | ||
</gallery> | </gallery> | ||
| Line 28: | Line 29: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | Our approach for using the landmarks is to manually mark the distinct anatomical features by point landmarks after the automatic registration is done. Matching of landmarks is performed by two different | + | Our approach for using the landmarks is to manually mark the distinct anatomical features by point landmarks after the automatic registration is done. Matching of landmarks is performed by two different methods. In one the optimization procedure is applied to the cost function containing an intensity-based data term and a landmark term, sum of squares of distances between fixed and moving landmarks. In another, an initial deformation field obtained from automatic registration is corrected by the field built on RBFs around the landmarks. |
Our plan for the project week is to first try out <bar>,... | Our plan for the project week is to first try out <bar>,... | ||
| Line 37: | Line 38: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | The code for the two | + | The code for the two methods of landmark matching has been implemented to our in house open source Plastimatch software. The methods have been validated on synthetic images and on lung CT scans obtained from NBIA data base. Statistical analysis for method validation has been performed on 4D CT scans obtained from http://www.dir-lab.com |
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 19:11, 6 June 2010
Home < 2010 Summer Project Week Fiducial Deformable Registration
Key Investigators
- MGH: Nadya Shusharina, Greg Sharp
Objective
We are developing methods for deformable image registration with landmark information. The goal is to improve the registration if misaligned areas are found.
Approach, Plan
Our approach for using the landmarks is to manually mark the distinct anatomical features by point landmarks after the automatic registration is done. Matching of landmarks is performed by two different methods. In one the optimization procedure is applied to the cost function containing an intensity-based data term and a landmark term, sum of squares of distances between fixed and moving landmarks. In another, an initial deformation field obtained from automatic registration is corrected by the field built on RBFs around the landmarks.
Our plan for the project week is to first try out <bar>,...
Progress
The code for the two methods of landmark matching has been implemented to our in house open source Plastimatch software. The methods have been validated on synthetic images and on lung CT scans obtained from NBIA data base. Statistical analysis for method validation has been performed on 4D CT scans obtained from http://www.dir-lab.com
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below)
- ITK Module
- Slicer Module
- Built-in
- Extension -- commandline
- Extension -- loadable
- Other (YES) -- stand alone utility
References
- G. C. Sharp, R. Li, J. Wolfgang, G. TY Chen, M. Peroni, M. F. Spadea, S. Mori, J. Zhang, J. Shackleford, N. Kandasamy, Plastimatch - an open source software suite for radiotherapy image processing, In the proceedings of the 16th International Conference on the Use of Computers in Radiotherapy (ICCR 2010), Amsterdam, The Netherlands.
- Plastimatch. http://plastimatch.org