Difference between revisions of "2010 Winter Project Week LongitudinalLupusAnalyses"
From NAMIC Wiki
Hjbockholt (talk | contribs) |
(Added images) |
||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-SLC2010.png|[[2010_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2010.png|[[2010_Winter_Project_Week#Projects|Projects List]] | ||
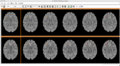
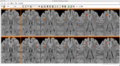
| − | Image: | + | Image:CompareViewFlairLesionDiffWholeBrain.png|Flair with lesion diff. |
| − | Image: | + | Image:CompareViewFlairLesionDiff.png|Lesions Time1 Vs. Time2 |
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * MRN: Mark Scully, Jeremy Bockholt |
| − | * | + | * BWH: Steve Pieper |
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 14: | Line 14: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | |||
| + | Improve support for longitudinal analysis of lesion data. | ||
| Line 27: | Line 27: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | '''1.''' Meet with Andriy Fedorov and go over existing Change Tracker codebase and determine whether it can be adapted and if not, what elements can be reused. | |
| − | + | '''2.''' Incorporate our current approach into Change Tracker or Re-Use elements of Change Tracker in a new longitudinal lesion analysis tool. | |
</div> | </div> | ||
| Line 36: | Line 36: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | ||
| + | '''1.''' Created a pipeline for longitudinal segmentation. | ||
| + | |||
| + | '''2.''' Created a module to produce lesion change maps. | ||
| + | |||
| + | |||
| Line 45: | Line 50: | ||
==References== | ==References== | ||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
Latest revision as of 15:40, 8 January 2010
Home < 2010 Winter Project Week LongitudinalLupusAnalysesKey Investigators
- MRN: Mark Scully, Jeremy Bockholt
- BWH: Steve Pieper
Objective
Improve support for longitudinal analysis of lesion data.
Approach, Plan
1. Meet with Andriy Fedorov and go over existing Change Tracker codebase and determine whether it can be adapted and if not, what elements can be reused.
2. Incorporate our current approach into Change Tracker or Re-Use elements of Change Tracker in a new longitudinal lesion analysis tool.
Progress
1. Created a pipeline for longitudinal segmentation.
2. Created a module to produce lesion change maps.