Difference between revisions of "2010 Winter Project Week VervetMRILongitudinalAnalysis"
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | <gallery perrow=" | + | <gallery perrow="4"> |
Image:PW-SLC2010.png|[[2010_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2010.png|[[2010_Winter_Project_Week#Projects|Projects List]] | ||
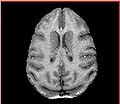
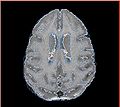
| − | Image: | + | Image:Calvin_atlas_demo.jpg | Axial slice for one of the vervet subjects |
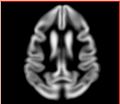
| − | Image: | + | Image:Calvin_atlas_demo_rhesus_gm.jpg | Aligned rhesus grey matter probabilistic atlas |
| − | Image: | + | Image:Calvin_atlas_demo_vervet_gm.jpg | Aligned vervet grey matter probabilistic atlas |
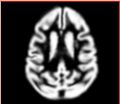
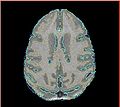
| − | Image: | + | Image:Calvin_atlas_demo_segm.jpg | Overlayed segmentation result by EM Segmenter with vervet probabilistic atlas |
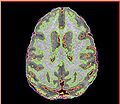
| + | Image:Calvin_ABC.jpg | Segmentation results by ABC | ||
| + | Image:Calvin_EMSegment.jpg | Segmentation result by EMSegment in Slicer 2.8 | ||
</gallery> | </gallery> | ||
| Line 13: | Line 15: | ||
* Andriy Fedorov, Ron Kikinis (BWH) | * Andriy Fedorov, Ron Kikinis (BWH) | ||
* Ginger Li, Chris Wyatt (VT) | * Ginger Li, Chris Wyatt (VT) | ||
| + | * Martin Styner (UNC) | ||
| + | * Kilian Pohl (IBM) | ||
| + | * Hans Johnson (UIowa) | ||
| + | * Marcel Prastawa (SCI) | ||
| Line 33: | Line 39: | ||
'''2.''' Build probabilistic tissue atlas using the baseline images | '''2.''' Build probabilistic tissue atlas using the baseline images | ||
| − | '''3.''' Use the atlas and automated segmentation tools to identify subcortical structures in all subjects for the baseline and follow-up | + | '''3.''' Register follow-up to baseline |
| + | |||
| + | '''4.''' Use the atlas and automated segmentation tools to identify subcortical structures in all subjects for the baseline and follow-up | ||
Alternatively, instead of the Steps 1 and 2 we could use UNC Rhesus atlas, if the structural changes between the rhesus and vervet brains are not significant, and we can successfully perform deformable registration. | Alternatively, instead of the Steps 1 and 2 we could use UNC Rhesus atlas, if the structural changes between the rhesus and vervet brains are not significant, and we can successfully perform deformable registration. | ||
| − | + | The focus of our work at AHM will be on Step 4 above. Given two types of atlases: | |
| + | |||
| + | * Vervet atlas we constructed | ||
| + | * Rhesus atlas prepared by UNC group | ||
| + | |||
| + | ... and three approaches to segmentation: | ||
| + | |||
| + | * EM Segmenter in Slicer 3.4 | ||
| + | * itkEMS and/or ABC Slicer 3.5 extension (are they different?) | ||
| + | * fluid deformable registration of the atlas to a subject | ||
| + | |||
| + | we will study what combination works best for our data, based on the subjective evaluation of the segmentation results. | ||
</div> | </div> | ||
| Line 48: | Line 67: | ||
* prepared 3-class and subcortical segmentations for one subject (semi-manually) | * prepared 3-class and subcortical segmentations for one subject (semi-manually) | ||
* atlas construction workflow is now composed completely from Slicer/NA-MIC Kit tools | * atlas construction workflow is now composed completely from Slicer/NA-MIC Kit tools | ||
| − | * initial version of the vervet probabilistic atlas completed | + | * initial version of the re-worked vervet probabilistic atlas completed |
''During the meeting'': | ''During the meeting'': | ||
| − | * | + | * discussed registration with Hans, Martin; looked at atlas to subject registration, confirmed accuracy is good for segmentation |
| − | * | + | * Worked with Marcel / ABC segmentation module -- worked out of the box, very few parameters need to be specified. Went through couple of iterations to expose some parameters of ABC (arbitrary number of classes, allow to disable registration, allow to specify priors per class). |
| − | * | + | * Worked with Kilian / EMSegmenter. Demo of Slicer2 EMSegmenter capabilities, not available in Slicer3. Fine-tuned parameters to achieve very good 3-class segmentation results on one subject. |
| + | * Worked with Daniel / VMTK segmentation to remove vessels prior to segmentation. | ||
| + | * Discussed the use of additional modalities for segmentation (T2 & PD). Agreed resolution not sufficient to improve results (4 times slice thickness). Single-shot T2/PD DICOM reading issue with Slicer nrrd reader: reported to Xiaodong | ||
| + | |||
</div> | </div> | ||
| Line 62: | Line 84: | ||
==References== | ==References== | ||
| − | * NA-MIC | + | * [[NA-MIC_NCBC_Collaboration:Measuring_Alcohol_and_Stress_Interaction|NA-MIC NCBC Collaboration:Measuring Alcohol and Stress Interaction]] |
| − | * | + | * [[October_08_2009_Meeting:_VT-NAMIC_Revised_atlas_construction_workflow|Vervet atlas construction workflow]] |
| − | * Relevant papers and studies from UNC | + | * Relevant papers and studies from UNC: |
| − | * BRAINSFit registration | + | ** [[DBP2:UNC:Local_Cortical_Thickness_Pipeline#Brain_atlases|UNC Rhesus brain atlas]] |
| − | * N4 bias correction | + | ** [[DBP2:UNC:Cortical_Thickness_Roadmap|Comparison between EM Segmenter and itkEMS]] |
| − | * EM Segmenter | + | ** M. Styner, R. Knickmeyer, S. Joshi, C. Coe, S. J. Short, and J. Gilmore. Automatic brain segmentation in rhesus monkeys. Proc SPIE Medical Imaging Conference, Proc SPIE Vol 6512 Medical Imaging 2007, pp 65122L-1 - 65122L-8 [http://www.google.com/url?sa=t&source=web&ct=res&cd=1&url=http%3A%2F%2Fwww.cs.unc.edu%2F~styner%2Fpublic%2FMyPapers%2Fpapers%2FMedical%2520Imaging%25202007%2520Image%2520Processing%25202007%2520Styner- |
| − | * itkEMS | + | ** R. C. Knickmeyer, M. Styner, S. J. Short, G. R. Lubach, C. Kang, R. Hamer, C. L. Coe, and J. H. Gilmore, "Maturational trajectories of cortical brain development through the pubertal transition: Unique species and sex differences in the monkey revealed through structural magnetic resonance imaging," pp. bhp166+, August 2009. [http://dx.doi.org/10.1093/cercor/bhp166 DOI] |
| − | * . | + | * [http://www.nitrc.org/projects/multimodereg/ BRAINSFit registration tools (Hans Johnson, UIowa) on NITRC] |
| + | * [http://www.insight-journal.org/browse/publication/640 N4 bias correction (Nick Tustison, UPenn) on Insight Journal] | ||
| + | * [http://wiki.slicer.org/slicerWiki/index.php/Modules:EMSegment-TemplateBuilder Slicer 3.4 EM Segmenter] | ||
| + | * [http://www.ia.unc.edu/dev/download/itkems/index.htm UNC itkEMS] | ||
| + | * Slicer 3.5 ABC extension | ||
</div> | </div> | ||
Latest revision as of 17:34, 8 January 2010
Home < 2010 Winter Project Week VervetMRILongitudinalAnalysis
Key Investigators
- Andriy Fedorov, Ron Kikinis (BWH)
- Ginger Li, Chris Wyatt (VT)
- Martin Styner (UNC)
- Kilian Pohl (IBM)
- Hans Johnson (UIowa)
- Marcel Prastawa (SCI)
Objective
We are developing methods for analysis of structural images from nonhuman primates for identifying regional differences in brain morphometry caused by interactions of stress and alcohol exposure. Specifically, we are focused on vervet monkey species. Our objective is to develop image analysis workflow for accurate and reproducible segmentation of vervet brain to facilitate morphometry studies. Ideally, we would like to have the complete workflow implemented using 3DSlicer tools.
Approach, Plan
Given two serial T1 scans for 10 vervet subjects, our approach is the following:
1. (Semi-)manually segment GM/WM/CSF and certain subcortical structures for one of the subjects
2. Build probabilistic tissue atlas using the baseline images
3. Register follow-up to baseline
4. Use the atlas and automated segmentation tools to identify subcortical structures in all subjects for the baseline and follow-up
Alternatively, instead of the Steps 1 and 2 we could use UNC Rhesus atlas, if the structural changes between the rhesus and vervet brains are not significant, and we can successfully perform deformable registration.
The focus of our work at AHM will be on Step 4 above. Given two types of atlases:
- Vervet atlas we constructed
- Rhesus atlas prepared by UNC group
... and three approaches to segmentation:
- EM Segmenter in Slicer 3.4
- itkEMS and/or ABC Slicer 3.5 extension (are they different?)
- fluid deformable registration of the atlas to a subject
we will study what combination works best for our data, based on the subjective evaluation of the segmentation results.
Progress
Prior to the meeting (current progress and notes):
- completely redesigned atlas construction workflow, prepared detailed documentation
- prepared 3-class and subcortical segmentations for one subject (semi-manually)
- atlas construction workflow is now composed completely from Slicer/NA-MIC Kit tools
- initial version of the re-worked vervet probabilistic atlas completed
During the meeting:
- discussed registration with Hans, Martin; looked at atlas to subject registration, confirmed accuracy is good for segmentation
- Worked with Marcel / ABC segmentation module -- worked out of the box, very few parameters need to be specified. Went through couple of iterations to expose some parameters of ABC (arbitrary number of classes, allow to disable registration, allow to specify priors per class).
- Worked with Kilian / EMSegmenter. Demo of Slicer2 EMSegmenter capabilities, not available in Slicer3. Fine-tuned parameters to achieve very good 3-class segmentation results on one subject.
- Worked with Daniel / VMTK segmentation to remove vessels prior to segmentation.
- Discussed the use of additional modalities for segmentation (T2 & PD). Agreed resolution not sufficient to improve results (4 times slice thickness). Single-shot T2/PD DICOM reading issue with Slicer nrrd reader: reported to Xiaodong
References
- NA-MIC NCBC Collaboration:Measuring Alcohol and Stress Interaction
- Vervet atlas construction workflow
- Relevant papers and studies from UNC:
- UNC Rhesus brain atlas
- Comparison between EM Segmenter and itkEMS
- M. Styner, R. Knickmeyer, S. Joshi, C. Coe, S. J. Short, and J. Gilmore. Automatic brain segmentation in rhesus monkeys. Proc SPIE Medical Imaging Conference, Proc SPIE Vol 6512 Medical Imaging 2007, pp 65122L-1 - 65122L-8 [http://www.google.com/url?sa=t&source=web&ct=res&cd=1&url=http%3A%2F%2Fwww.cs.unc.edu%2F~styner%2Fpublic%2FMyPapers%2Fpapers%2FMedical%2520Imaging%25202007%2520Image%2520Processing%25202007%2520Styner-
- R. C. Knickmeyer, M. Styner, S. J. Short, G. R. Lubach, C. Kang, R. Hamer, C. L. Coe, and J. H. Gilmore, "Maturational trajectories of cortical brain development through the pubertal transition: Unique species and sex differences in the monkey revealed through structural magnetic resonance imaging," pp. bhp166+, August 2009. DOI
- BRAINSFit registration tools (Hans Johnson, UIowa) on NITRC
- N4 bias correction (Nick Tustison, UPenn) on Insight Journal
- Slicer 3.4 EM Segmenter
- UNC itkEMS
- Slicer 3.5 ABC extension