Difference between revisions of "2010 Winter Project Week VervetMRILongitudinalAnalysis"
| Line 53: | Line 53: | ||
''During the meeting'': | ''During the meeting'': | ||
| − | * testing and fine-tuning segmentation tools | + | * testing and fine-tuning segmentation tools -- ''Work with: EM Segmenter/Kilian Pohl, itkEMS/ABC/Marcel Prastawa''; try to complete segmentation of baseline and followup scans |
| − | * improving deformable registration of rhesus atlas to vervet ([[October_08_2009_Meeting:_VT-NAMIC_Revised_atlas_construction_workflow#Template_Gucci |initial results available]]) | + | * improving deformable registration of rhesus atlas to vervet ([[October_08_2009_Meeting:_VT-NAMIC_Revised_atlas_construction_workflow#Template_Gucci |initial results available]]) ''Work with: Luis Ibanez, Jim Miller, Dominik Maier, Casey Goodlett, Hans Johnson ... any expertise in registration is welcome'' |
* collect overall comments on the workflow from the related experience in similar workflows (Martin Styner) | * collect overall comments on the workflow from the related experience in similar workflows (Martin Styner) | ||
Revision as of 00:45, 12 December 2009
Home < 2010 Winter Project Week VervetMRILongitudinalAnalysis
Key Investigators
- Andriy Fedorov, Ron Kikinis (BWH)
- Ginger Li, Chris Wyatt (VT)
Objective
We are developing methods for analysis of structural images from nonhuman primates for identifying regional differences in brain morphometry caused by interactions of stress and alcohol exposure. Specifically, we are focused on vervet monkey species. Our objective is to develop image analysis workflow for accurate and reproducible segmentation of vervet brain to facilitate morphometry studies. Ideally, we would like to have the complete workflow implemented using 3DSlicer tools.
Approach, Plan
Given two serial T1 scans for 10 vervet subjects, our approach is the following:
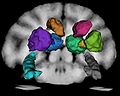
1. (Semi-)manually segment GM/WM/CSF and certain subcortical structures for one of the subjects
2. Build probabilistic tissue atlas using the baseline images
3. Register follow-up to baseline
4. Use the atlas and automated segmentation tools to identify subcortical structures in all subjects for the baseline and follow-up
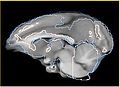
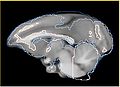
Alternatively, instead of the Steps 1 and 2 we could use UNC Rhesus atlas, if the structural changes between the rhesus and vervet brains are not significant, and we can successfully perform deformable registration.
Question to EM experts: how good should be the alignment between atlas and subject for segmentation?
Progress
Prior to the meeting (current progress and notes):
- completely redesigned atlas construction workflow, prepared detailed documentation
- prepared 3-class and subcortical segmentations for one subject (semi-manually)
- atlas construction workflow is now composed completely from Slicer/NA-MIC Kit tools
- initial version of the vervet probabilistic atlas completed
During the meeting:
- testing and fine-tuning segmentation tools -- Work with: EM Segmenter/Kilian Pohl, itkEMS/ABC/Marcel Prastawa; try to complete segmentation of baseline and followup scans
- improving deformable registration of rhesus atlas to vervet (initial results available) Work with: Luis Ibanez, Jim Miller, Dominik Maier, Casey Goodlett, Hans Johnson ... any expertise in registration is welcome
- collect overall comments on the workflow from the related experience in similar workflows (Martin Styner)
References
- NA-MIC collaboration page
- Workflow/progress page
- Relevant papers and studies from UNC
- BRAINSFit registration
- N4 bias correction
- EM Segmenter
- itkEMS
- ...