Difference between revisions of "2010 Winter Project Week WM ATLAS"
| (29 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-SLC2010.png|[[2010_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2010.png|[[2010_Winter_Project_Week#Projects|Projects List]] | ||
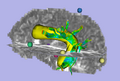
| − | Image:WM-seg-1.png|Automatically segmented fiber tracts. | + | Image:WM-seg-1.png|Input from example subject: Automatically segmented fiber tracts and fMRI peak locations. This data from 5 subjects has been used in matlab to build a fiber-fMRI distance model to enable prediction/detection of fiber tracts using fMRI. |
| − | Image: | + | Image:SlicerImage_1.png|Output: Yellow region is prediction of arcuate location in this patient, based on fMRI peaks (spheres). Predicted AF visualized in Slicer3 with fMRI points and actual patient's fiber tracts. |
| + | Image:SlicerImage_0.png|Output: Predicted AF location visualized in Slicer3 with fibers clustered by python test code and colored using Mahnaz Maddah's cluster coloring in Slicer3. | ||
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * Lauren O'Donnell ( | + | * Lauren O'Donnell (Harvard Medical School (BWH)) |
| + | * Luis Ibanez (Kitware Inc) | ||
* Carl-Fredrik Westin (Professor, Harvard Medical School (BWH) ) | * Carl-Fredrik Westin (Professor, Harvard Medical School (BWH) ) | ||
* William Wells (Professor, Harvard Medical School (BWH) ) | * William Wells (Professor, Harvard Medical School (BWH) ) | ||
* Alexandra J Golby (Professor of Neurosurgery, Harvard Medical School (BWH) ) | * Alexandra J Golby (Professor of Neurosurgery, Harvard Medical School (BWH) ) | ||
| + | * Gordon L. Kindlmann (Professor University of Chicago) (teem help/docs/implementation) | ||
| + | * James Malcolm and Yogesh Rathi (2-tensor tractography) | ||
| + | * Steve Pieper (advice on ipython and module implementation) | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 17: | Line 22: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | Implement tract-based morphometry pipeline in 3D Slicer, including atlas-based WM (white matter) segmentation and whole-brain WM coordinate system generation. Investigate integration of FMRI+DTI into atlas pipeline, for neurosurgical planning applications. | + | Eventual goal: Implement slicer module for neurosurgical planning using novel fMRI+DTI atlas. |
| + | |||
| + | Intermediate goals: Investigate re-implementation of tract-based morphometry pipeline in 3D Slicer, including clustering for atlas creation, atlas-based WM (white matter) segmentation and whole-brain WM coordinate system generation. Investigate integration of FMRI+DTI into atlas pipeline, for neurosurgical planning applications. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| Line 29: | Line 31: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | The original pipeline consists of (over a group): | + | The original WM atlas pipeline consists of (over a group): |
* Preprocessing | * Preprocessing | ||
| − | ** DTI and FA image calculation | + | ** DTI and FA image calculation (slicer) |
| − | ** congealing registration of FA images | + | ** congealing registration of FA images (Lilla's code) |
| − | ** whole-brain tract seeding | + | ** whole-brain tract seeding (slicer2) |
| − | ** application of registration to tractography | + | ** application of registration to tractography (matlab) |
| − | * | + | ** measurement of fiber-fMRI model (slicer2/matlab) |
| − | * | + | * Atlas generation (matlab) |
** clustering of (sample from) whole-brain tractography in group | ** clustering of (sample from) whole-brain tractography in group | ||
** expert labeling of clusters to give anatomical structures | ** expert labeling of clusters to give anatomical structures | ||
** the above produces an atlas that can be used to segment | ** the above produces an atlas that can be used to segment | ||
| − | * analysis of segmented WM | + | * WM tract segmentation (matlab) |
| + | ** based on atlas, plus fMRI in future for IGT | ||
| + | * analysis of segmented WM (matlab) | ||
** optimal generation of tract-based coordinate systems per anatomical structure | ** optimal generation of tract-based coordinate systems per anatomical structure | ||
** export of cluster/structure based measurements (FA, etc) | ** export of cluster/structure based measurements (FA, etc) | ||
** some statistical analysis | ** some statistical analysis | ||
| − | + | ||
| − | + | We propose re-implementation of much of the above in python and/or Slicer3 framework. This will enable use of the pipeline by others and our investigation of DTI+fMRI atlases for neurosurgery. | |
| − | |||
</div> | </div> | ||
| Line 53: | Line 56: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | We have tested/debugged the group registration and enabled Slicer3 display of matlab output for tract prediction. | ||
| + | |||
* Preprocessing | * Preprocessing | ||
| − | ** | + | ** Debugging of ITK congealing with Luis. |
| − | |||
** implementation of initial seeding pipeline script in ipython | ** implementation of initial seeding pipeline script in ipython | ||
| − | + | ** Gordon has implemented tractography methods and some probing capability in teem, may allow more direct teem/itk/vtk dependencies | |
| − | ** Gordon has implemented some probing capability in teem, may allow more direct teem/itk/vtk dependencies | + | * Clustering |
| − | ** | + | ** initial single-subject cluster test implemented in python |
| + | ** communicated with Mahnaz Maddah to learn how to display clusters in Slicer3 (cell data array must be called ClusterId) | ||
| + | * IGT fMRI+DTI atlas | ||
| + | ** Initial methods development: fiber-fMRI distance model (MICCAI Diffusion workshop 2009) | ||
| + | ** Output of matlab-generated tract predictions to slicer3 as nrrd volumes | ||
</div> | </div> | ||
</div> | </div> | ||
<div style="width: 97%; float: left;"> | <div style="width: 97%; float: left;"> | ||
| + | |||
| + | ==References== | ||
| + | |||
| + | This work is based on the papers | ||
| + | |||
| + | Defining Spatial Relationships Between fMRI and DTI Fiber Tracts | ||
| + | Lauren J. O'Donnell, Laura E. Rigolo, Isaiah Norton, Carl-Fredrik Westin, and Alexandra J. Golby. | ||
| + | MICCAI Workshop on Diffusion Modeling and the Fibre Cup, 2009 | ||
| + | |||
| + | Tract-Based Morphometry for White Matter Group Analysis. Lauren J. O'Donnell, Carl-Fredrik Westin, and Alexandra J. Golby. | ||
| + | NeuroImage 45(3):832-844, 2009 | ||
| + | |||
| + | Automatic Tractography Segmentation Using a High-Dimensional White Matter Atlas. | ||
| + | Lauren O'Donnell and Carl-Fredrik Westin. | ||
| + | IEEE Transactions in Medical Imaging 26(11):1562-1575, 2007 | ||
| + | </div> | ||
Latest revision as of 17:42, 8 January 2010
Home < 2010 Winter Project Week WM ATLASKey Investigators
- Lauren O'Donnell (Harvard Medical School (BWH))
- Luis Ibanez (Kitware Inc)
- Carl-Fredrik Westin (Professor, Harvard Medical School (BWH) )
- William Wells (Professor, Harvard Medical School (BWH) )
- Alexandra J Golby (Professor of Neurosurgery, Harvard Medical School (BWH) )
- Gordon L. Kindlmann (Professor University of Chicago) (teem help/docs/implementation)
- James Malcolm and Yogesh Rathi (2-tensor tractography)
- Steve Pieper (advice on ipython and module implementation)
Objective
Eventual goal: Implement slicer module for neurosurgical planning using novel fMRI+DTI atlas.
Intermediate goals: Investigate re-implementation of tract-based morphometry pipeline in 3D Slicer, including clustering for atlas creation, atlas-based WM (white matter) segmentation and whole-brain WM coordinate system generation. Investigate integration of FMRI+DTI into atlas pipeline, for neurosurgical planning applications.
Approach, Plan
The original WM atlas pipeline consists of (over a group):
- Preprocessing
- DTI and FA image calculation (slicer)
- congealing registration of FA images (Lilla's code)
- whole-brain tract seeding (slicer2)
- application of registration to tractography (matlab)
- measurement of fiber-fMRI model (slicer2/matlab)
- Atlas generation (matlab)
- clustering of (sample from) whole-brain tractography in group
- expert labeling of clusters to give anatomical structures
- the above produces an atlas that can be used to segment
- WM tract segmentation (matlab)
- based on atlas, plus fMRI in future for IGT
- analysis of segmented WM (matlab)
- optimal generation of tract-based coordinate systems per anatomical structure
- export of cluster/structure based measurements (FA, etc)
- some statistical analysis
We propose re-implementation of much of the above in python and/or Slicer3 framework. This will enable use of the pipeline by others and our investigation of DTI+fMRI atlases for neurosurgery.
Progress
We have tested/debugged the group registration and enabled Slicer3 display of matlab output for tract prediction.
- Preprocessing
- Debugging of ITK congealing with Luis.
- implementation of initial seeding pipeline script in ipython
- Gordon has implemented tractography methods and some probing capability in teem, may allow more direct teem/itk/vtk dependencies
- Clustering
- initial single-subject cluster test implemented in python
- communicated with Mahnaz Maddah to learn how to display clusters in Slicer3 (cell data array must be called ClusterId)
- IGT fMRI+DTI atlas
- Initial methods development: fiber-fMRI distance model (MICCAI Diffusion workshop 2009)
- Output of matlab-generated tract predictions to slicer3 as nrrd volumes
References
This work is based on the papers
Defining Spatial Relationships Between fMRI and DTI Fiber Tracts Lauren J. O'Donnell, Laura E. Rigolo, Isaiah Norton, Carl-Fredrik Westin, and Alexandra J. Golby. MICCAI Workshop on Diffusion Modeling and the Fibre Cup, 2009
Tract-Based Morphometry for White Matter Group Analysis. Lauren J. O'Donnell, Carl-Fredrik Westin, and Alexandra J. Golby. NeuroImage 45(3):832-844, 2009
Automatic Tractography Segmentation Using a High-Dimensional White Matter Atlas. Lauren O'Donnell and Carl-Fredrik Westin. IEEE Transactions in Medical Imaging 26(11):1562-1575, 2007