Difference between revisions of "2011 Winter Project Week:GAMBITCorticalThicknessAnalysis"
From NAMIC Wiki
(Created page with '__NOTOC__ <gallery> Image:PW-SLC2011.png|Projects List Image:Arctic_Logo.png|[http://www.nitrc.org/projects/arctic ARCTIC: Automatic Regi…') |
|||
| (7 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
Image:PW-SLC2011.png|[[2011_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2011.png|[[2011_Winter_Project_Week#Projects|Projects List]] | ||
Image:Arctic_Logo.png|[http://www.nitrc.org/projects/arctic ARCTIC: Automatic Regional Cortical ThICkness] | Image:Arctic_Logo.png|[http://www.nitrc.org/projects/arctic ARCTIC: Automatic Regional Cortical ThICkness] | ||
| − | Image: | + | Image:GAMBITLogo.png|[http://www.nitrc.org/projects/gambit GAMBIT: Group-wise Automatic Mesh-Based analysis of cortIcal Thickness] |
| − | Image: | + | Image:BrainDevelopment.jpg|Brain development |
</gallery> | </gallery> | ||
{| | {| | ||
| − | |||
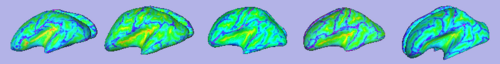
| [[Image:GAMBIT_QC_SulcalDepth.png|thumb|500px|Sulcal depth overlayed on inflated cortical surfaces]] | | [[Image:GAMBIT_QC_SulcalDepth.png|thumb|500px|Sulcal depth overlayed on inflated cortical surfaces]] | ||
|} | |} | ||
| Line 43: | Line 42: | ||
*[http://www.nitrc.org/projects/gambit GAMBIT: Group-wise Automatic Mesh-Based analysis of cortIcal Thickness ] | *[http://www.nitrc.org/projects/gambit GAMBIT: Group-wise Automatic Mesh-Based analysis of cortIcal Thickness ] | ||
| − | Our plan for the project week is to | + | Our plan for the project week is to improve GAMBIT's overall distribution: |
| − | + | *Include/Improve download/compilation of external packages | |
| − | + | *Improve ShapeWorks integration within Slicer3 (meeting with SCI members) | |
| − | |||
</div> | </div> | ||
| Line 54: | Line 52: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | *Attended the Particle Shape Correspondence Breakout | |
| + | ** ShapeWorks will be available soon as a SVN repository on NITRC (from CVS on GForge) and directly integrated within 3D Slicer as an extension | ||
| + | *GAMBIT now includes download/compilation of external packages: ABC, ShapeWorks, BRAINSFit, BRAINSResample, BRAINSDemonWarp... | ||
</div> | </div> | ||
Latest revision as of 17:35, 14 January 2011
Home < 2011 Winter Project Week:GAMBITCorticalThicknessAnalysisKey Investigators
- UNC: Clement Vachet, Heather Cody Hazlett, Martin Styner
Objective
We would like to create end-to-end applications within Slicer3 allowing voxel-based and mesh-based group analyses of cortical thickness.
Such a workflow applied to the young brain (2-4 years old) is our goal in order to start a longitudinal study of early brain development in autism (UNC DBP).
See our Roadmap for more details.
Approach, Plan
Our approach is described in the following wiki pages:
Two 3D Slicer high-level modules -directly available as extensions- have been developped in that regard:
- ARCTIC: Automatic Regional Cortical ThICkness
- GAMBIT: Group-wise Automatic Mesh-Based analysis of cortIcal Thickness
Our plan for the project week is to improve GAMBIT's overall distribution:
- Include/Improve download/compilation of external packages
- Improve ShapeWorks integration within Slicer3 (meeting with SCI members)
Progress
- Attended the Particle Shape Correspondence Breakout
- ShapeWorks will be available soon as a SVN repository on NITRC (from CVS on GForge) and directly integrated within 3D Slicer as an extension
- GAMBIT now includes download/compilation of external packages: ABC, ShapeWorks, BRAINSFit, BRAINSResample, BRAINSDemonWarp...
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a:
- ITK Module
- Slicer Module
- Built-in
- Extension -- commandline - YES
- Extension -- loadable
- Other (Please specify)
References
- C. Vachet, H. Cody Hazlett, M. Niethammer, I. Oguz, J. Cates, R. Whitaker, J. Piven, M. Styner, Group-wise Automatic Mesh-Based Analysis of Cortical Thickness, accepted to SPIE 2011
- I. Oguz, M. Niethammer, J. Cates, R. Whitaker, T. Fletcher, C. Vachet, and M. Styner, Cortical Correspondence with Probabilistic Fiber Connectivity, Information Processing in Medical Imaging, IPMI 2009, LNCS, in print.
- H.C. Hazlett, C. Vachet, C. Mathieu, M. Styner, J. Piven, Use of the Slicer3 Toolkit to Produce Regional Cortical Thickness Measurement of Pediatric MRI Data, presented at the 8th Annual International Meeting for Autism Research (IMFAR) Chicago, IL 2009.
- C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Mesh-based Local Cortical Thickness Framework, UNC Radiology Research Day 2010 abstract
- C. Mathieu, C. Vachet, H.C. Hazlett, G. Geric, J. Piven, and M. Styner, ARCTIC – Automatic Regional Cortical ThICkness Tool, UNC Radiology Research Day 2009 abstract
- J Cates, P T Fletcher, M Styner, M Shenton, R Whitaker. Shape Modeling and Analysis with Entropy-Based Particle Systems. Information Processing in Medical Imaging IPMI 2007, LNCS 4584, pp. 333-345, 2007