Difference between revisions of "2011 Winter Project Week:MRSI module and SIVIC interface"
| (11 intermediate revisions by 3 users not shown) | |||
| Line 5: | Line 5: | ||
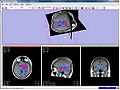
Image:Mrsi parametric-map-NAA 3d.jpg|Figure 2 - MRSI metabolite maps visualized in Slicer | Image:Mrsi parametric-map-NAA 3d.jpg|Figure 2 - MRSI metabolite maps visualized in Slicer | ||
Image:Mrsi_slicer_sivic.jpg| Figure 3 - Screenshot of the SIVIC MRSI module integrated into Slicer. | Image:Mrsi_slicer_sivic.jpg| Figure 3 - Screenshot of the SIVIC MRSI module integrated into Slicer. | ||
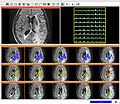
| + | Image:Sivic_slicer_slc_mets.jpg| Figure 4 - MRSI metabolite maps generated in SIVIC module referenced to anatomical (FLAIR) image. | ||
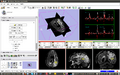
| + | Image:SIVICSlicerMultitrace.png| Figure 5 - Multiple trace functionality added to svk (SIVIC kit) during project week and then made accessible through the SIVIC MRSI Module. | ||
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
* MIT: Bjoern Menze, Mangpo Phothilimthana, Polina Golland | * MIT: Bjoern Menze, Mangpo Phothilimthana, Polina Golland | ||
| − | * UCSF: Olson | + | * UCSF: Beck Olson, Jason Crane, Sarah Nelson ([http://www.radiology.ucsf.edu/nelsonlab/ Nelson Lab]) |
| + | * BWH: Nicole Aucoin | ||
==Project== | ==Project== | ||
| Line 26: | Line 29: | ||
We want to integrate the [http://wiki.na-mic.org/Wiki/index.php/2010_WinterProject_Week_MRSIModule currently] available MRSI quantitation routines with the [http://sourceforge.net/apps/trac/sivic/ SIVIC] Slicer interface developed at the UCSF. | We want to integrate the [http://wiki.na-mic.org/Wiki/index.php/2010_WinterProject_Week_MRSIModule currently] available MRSI quantitation routines with the [http://sourceforge.net/apps/trac/sivic/ SIVIC] Slicer interface developed at the UCSF. | ||
| − | More specifically we want to | + | More specifically we want to integrate the previously developed signal processing routines (Fig. 1) into the svk library of the SIVIC framework, and realize the joint display of metabolic maps (Fig. 2) and spectral information (Fig. 3) within the current prototype of the Slicer module. |
</div> | </div> | ||
| Line 32: | Line 35: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
</div> | </div> | ||
| + | * We implemented a basic quantification algorithm (Fig. 4) integrating over predefined spectral regions and made progress towards implementing the other fitting routines. | ||
| + | |||
| + | * Metabolic maps generated in Slicer can now be displayed (Fig. 4). | ||
| + | |||
| + | * The display of individual spectra in Slicer now also supports multiple traces from the spectral fitting (Fig. 5). | ||
| + | |||
| + | * We improved the integration of SIVIC with Slicer through a better use of the MRML architecture. | ||
| + | |||
| + | * The new functionality is now available through SIVIC and a Slicer Module prototype. | ||
</div> | </div> | ||
| Line 46: | Line 58: | ||
##Built-in | ##Built-in | ||
##Extension -- commandline | ##Extension -- commandline | ||
| − | ##Extension -- loadable YES | + | ##Extension -- loadable YES |
| − | #Other (Please specify) | + | #Other (Please specify) YES: part of the 'vtk style' svk library of the SIVIC framework |
==References== | ==References== | ||
Latest revision as of 16:09, 14 January 2011
Home < 2011 Winter Project Week:MRSI module and SIVIC interfaceKey Investigators
- MIT: Bjoern Menze, Mangpo Phothilimthana, Polina Golland
- UCSF: Beck Olson, Jason Crane, Sarah Nelson (Nelson Lab)
- BWH: Nicole Aucoin
Project
Objective
Magnetic resonance spectroscopic imaging (MRSI) is a non-invasive diagnostic method used to determine the relative abundance of specific metabolites at arbitrary locations in vivo. Certain diseases -- such as tumors in brain, breast and prostate -- can be can be associated with characteristic changes in the metabolic level.
The objective of the current project is to develop a module proving the means for the processing and visualization of MRSI -- and thus for a joint analysis of magnetic resonance spectroscopic images together with other imaging modalities -- in Slicer.
Approach, Plan
Spectral fitting routines have been implemented, using a HSVD filter for water peak removal and baseline estimation, and a constrained non-linear least squares optimization for the fit of the resonance line models. Current fitting routines require, however, several external software libraries not to be distributed, installed and used easily.
We want to integrate the currently available MRSI quantitation routines with the SIVIC Slicer interface developed at the UCSF. More specifically we want to integrate the previously developed signal processing routines (Fig. 1) into the svk library of the SIVIC framework, and realize the joint display of metabolic maps (Fig. 2) and spectral information (Fig. 3) within the current prototype of the Slicer module.
Progress
- We implemented a basic quantification algorithm (Fig. 4) integrating over predefined spectral regions and made progress towards implementing the other fitting routines.
- Metabolic maps generated in Slicer can now be displayed (Fig. 4).
- The display of individual spectra in Slicer now also supports multiple traces from the spectral fitting (Fig. 5).
- We improved the integration of SIVIC with Slicer through a better use of the MRML architecture.
- The new functionality is now available through SIVIC and a Slicer Module prototype.
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below)
- ITK Module
- Slicer Module
- Built-in
- Extension -- commandline
- Extension -- loadable YES
- Other (Please specify) YES: part of the 'vtk style' svk library of the SIVIC framework
References
- Jason C. Crane, Marram P. Olson, Sarah J. Nelson. SIVIC: An Extensible Open-Source DICOM MR Spectroscopy Software Framework and Application Suite. Proc ISMRM 2010. 3354.