Difference between revisions of "2011 Winter Project Week:TwoTensorTracts"
From NAMIC Wiki
| (16 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-SLC2011.png|[[2011_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2011.png|[[2011_Winter_Project_Week#Projects|Projects List]] | ||
| − | Image: | + | Image:wholebrainclustersexample.jpg|Example whole brain clustering result. |
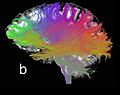
| − | Image: | + | Image:example2tensortracts.jpg|Example two-tensor tractography result. |
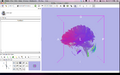
| + | Image:clustersinSlicer4.png|Progress: Cluster output loaded into Slicer4 with new MRML file. | ||
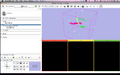
| + | Image:modelHierarchyClustersSlicer4.png|Progress: Model hierarchy created by initial Slicer4 module | ||
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
* BWH: Lauren O'Donnell, Yogesh Rathi, C-F Westin | * BWH: Lauren O'Donnell, Yogesh Rathi, C-F Westin | ||
| + | |||
| + | ==Thank You== | ||
| + | * BWH: Demian Wassermann (help with Python) | ||
| + | * Kitware: JC, J2, for bug squishing. Danielle Pace for pointers on Python modules for Slicer4. | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 28: | Line 27: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | Our | + | Our goals for the project week are to test 2-tensor tractography, to update clustering I/O to read tracts as a single large polydata file per subject, and to produce output in Slicer3 MRML format. |
| − | + | The next step is to ascertain what needs to be done to visualize/label the clusters in slicer3 or 4. This is in order to make atlases or study a particular tract. The old slicer2 interface allowed interactive selection of clusters (show/hide selected, and show/hide labeled or unlabeled) and anatomical naming/labeling. | |
</div> | </div> | ||
| Line 37: | Line 36: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | * Installed/tested 2-tensor slicer3 python module. Thank you Demian for help. Reported bug with voxel sizes to Yogesh Rathi. | |
| − | + | * Converted clustering output tcl script to output slicer3/slicer4 MRML format | |
| − | + | * Initial work on porting slicer2 ModelInteraction module to slicer4. Module will enable interactive model selection/grouping/naming. Slicer4 picking and improved model hierarchy functionality will be used when available. The module is a QTScriptedModule python Slicer4 implementation. | |
| + | * Some progress towards general file formats as input to clustering code, to allow slicer2/3/4/other programs to input tract files (one or many vtk's) | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 50: | Line 50: | ||
#ITK Module | #ITK Module | ||
| − | #Slicer Module | + | #Slicer Module YES |
| − | ##Built-in | + | ##Built-in YES! |
##Extension -- commandline | ##Extension -- commandline | ||
##Extension -- loadable | ##Extension -- loadable | ||
| Line 57: | Line 57: | ||
==References== | ==References== | ||
| − | * | + | *Tensor Kernels for Simultaneous Fiber Model Estimation and Tractography. Y. Rathi, J. G. Malcolm, O. Michailovich, C.-F. Westin, M. E. Shenton, S. Bouix. Magn Reson Med. 2010 |
| − | + | *Quantitative examination of a novel clustering method using magnetic resonance diffusion tensor tractography. A. N. Voineskos, L. J. O'Donnell, N. J. Lobaugh, D. Markant, S. H. Ameis, M. Niethammer, B. H. Mulsant, B. G. Pollock BG, J. L. Kennedy, C. F. Westin, M. E. Shenton. NeuroImage 45(2):370-376, 2009 | |
| − | + | *Automatic Tractography Segmentation Using a High-Dimensional White Matter Atlas. Lauren O'Donnell and Carl-Fredrik Westin. IEEE Transactions in Medical Imaging 26(11):1562-1575, 2007 | |
| − | * | ||
| − | |||
</div> | </div> | ||
Latest revision as of 18:07, 14 January 2011
Home < 2011 Winter Project Week:TwoTensorTractsKey Investigators
- BWH: Lauren O'Donnell, Yogesh Rathi, C-F Westin
Thank You
- BWH: Demian Wassermann (help with Python)
- Kitware: JC, J2, for bug squishing. Danielle Pace for pointers on Python modules for Slicer4.
Objective
To test 2-tensor tractography in Slicer3/standalone and hook it up to full-brain clustering pipelines.
Approach, Plan
Our goals for the project week are to test 2-tensor tractography, to update clustering I/O to read tracts as a single large polydata file per subject, and to produce output in Slicer3 MRML format.
The next step is to ascertain what needs to be done to visualize/label the clusters in slicer3 or 4. This is in order to make atlases or study a particular tract. The old slicer2 interface allowed interactive selection of clusters (show/hide selected, and show/hide labeled or unlabeled) and anatomical naming/labeling.
Progress
- Installed/tested 2-tensor slicer3 python module. Thank you Demian for help. Reported bug with voxel sizes to Yogesh Rathi.
- Converted clustering output tcl script to output slicer3/slicer4 MRML format
- Initial work on porting slicer2 ModelInteraction module to slicer4. Module will enable interactive model selection/grouping/naming. Slicer4 picking and improved model hierarchy functionality will be used when available. The module is a QTScriptedModule python Slicer4 implementation.
- Some progress towards general file formats as input to clustering code, to allow slicer2/3/4/other programs to input tract files (one or many vtk's)
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below)
- ITK Module
- Slicer Module YES
- Built-in YES!
- Extension -- commandline
- Extension -- loadable
- Other (Please specify)
References
- Tensor Kernels for Simultaneous Fiber Model Estimation and Tractography. Y. Rathi, J. G. Malcolm, O. Michailovich, C.-F. Westin, M. E. Shenton, S. Bouix. Magn Reson Med. 2010
- Quantitative examination of a novel clustering method using magnetic resonance diffusion tensor tractography. A. N. Voineskos, L. J. O'Donnell, N. J. Lobaugh, D. Markant, S. H. Ameis, M. Niethammer, B. H. Mulsant, B. G. Pollock BG, J. L. Kennedy, C. F. Westin, M. E. Shenton. NeuroImage 45(2):370-376, 2009
- Automatic Tractography Segmentation Using a High-Dimensional White Matter Atlas. Lauren O'Donnell and Carl-Fredrik Westin. IEEE Transactions in Medical Imaging 26(11):1562-1575, 2007