Difference between revisions of "2011 Winter Project Week: Python and Slicer4"
From NAMIC Wiki
| (12 intermediate revisions by 2 users not shown) | |||
| Line 8: | Line 8: | ||
* Jim - GE | * Jim - GE | ||
* Steve - Isomics | * Steve - Isomics | ||
| + | * Demian - BWH | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 34: | Line 35: | ||
** embed GUI in slicer interface (parent is slicer module panel) | ** embed GUI in slicer interface (parent is slicer module panel) | ||
** run as part of a workflow (parent is one step of workflow) | ** run as part of a workflow (parent is one step of workflow) | ||
| − | ** run GUI stand-alone (module's widget is top level window) | + | ** run GUI stand-alone (module's widget is top level window)- call this '''Slicelet''' mode. |
** run without GUI (parameters passed on command line CLI-style) | ** run without GUI (parameters passed on command line CLI-style) | ||
| Line 47: | Line 48: | ||
</div> | </div> | ||
| − | + | <div style="width: 97%; float: left;"> | |
==References== | ==References== | ||
| + | |||
| + | Example [http://viewvc.slicer.org/viewcvs.cgi/trunk/QTScriptedModules/Scripts/LabelStatistics.py?rev=15801&root=Slicer4&view=markup LabelStatistics module source code]. | ||
| + | |||
| + | {| | ||
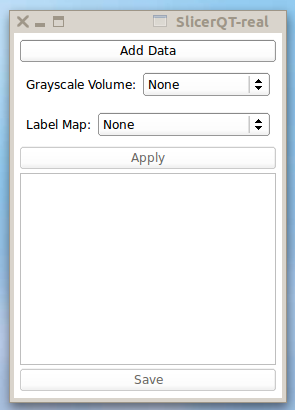
| + | |[[image:LabelStatistics-Slicelet-2011-01-09.png|thumb|Label Statistics running stand-alone as a Slicelet]]. | ||
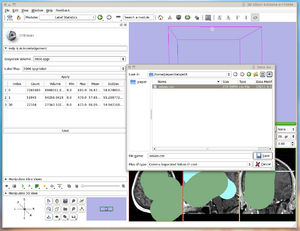
| + | |[[image:LabelStatistics-Module-2011-01-09.png|thumb|Label Statistics running as a slicer module]]. | ||
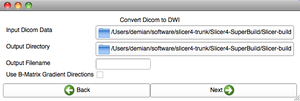
| + | |[[image:DWI2Tractography-Slicelet-2011-01-14.png|thumb|DWI to full brain tractography step 1]]. | ||
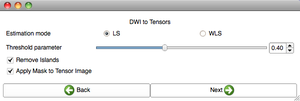
| + | |[[image:DWI2Tractography2-Slicelet-2011-01-14.png|thumb|DWI to full brain tractography step 2]]. | ||
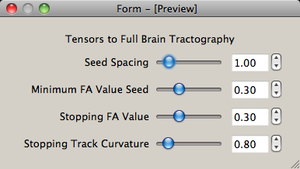
| + | |[[image:DWI2Tractography3-Slicelet-2011-01-14.png|thumb|DWI to full brain tractography step 3]]. | ||
| + | |} | ||
| + | |||
| + | |||
| + | Command line to run "Slicelet" from Slicer4 build directory: | ||
| + | |||
| + | <code> | ||
| + | % Slicer4 lib/Slicer3/QTScriptedModules/LabelStatistics.py | ||
| + | </code> | ||
| + | |||
| + | * TODO Items: | ||
| + | ** pass command line options to slicelets (use tclap-style --xml like the CLI modules) | ||
| + | ** optimize startup time for the interpreter | ||
| + | ** expose remaining parts of the Qt/qSlicer API to Python | ||
| + | ** progress and status output | ||
| + | ** wrapper to run non-scripted modules in slicelet mode | ||
| + | |||
| + | * Architectural Issues | ||
| + | ** Object inheritance plan for scripted modules and slicelets | ||
| + | ** Utilities needed to make it easy for developers/users to put together pipelines that mix module types | ||
| + | |||
</div> | </div> | ||
| − | |||
Latest revision as of 17:34, 14 January 2011
Home < 2011 Winter Project Week: Python and Slicer4Key Investigators
- Jc, Danielle - Kitware
- Jim - GE
- Steve - Isomics
- Demian - BWH
Objective
Develop scripting patterns for implementing workflows in slicer4 to port slicer3 functionality and streamline implementation of new functionality.
Approach, Plan
- Review initial experiences with PythonQt in slicer4
- Editor and SWidgets
- TubeTK-based Interactive PDF Segmenter
- Endoscopy
- Consider general architecture for creating module GUIs (scripted widgets)
- allow adaptive re-use of widgets in different modules such as:
- embed subset of painting tools inside interactive segmenters
- embed small model maker interface in segmentation tools
- embed some annotation (e.g. fiducial) for landmarking in registration
- embed CLI interfaces in workflow wizards
- define inheritance for PythonQt implementations to provide consistency across module GUI implementaitons
- allow adaptive re-use of widgets in different modules such as:
- Consider use cases for (scripted) slicer modules - can one implementation pattern cover all use cases?
- embed GUI in slicer interface (parent is slicer module panel)
- run as part of a workflow (parent is one step of workflow)
- run GUI stand-alone (module's widget is top level window)- call this Slicelet mode.
- run without GUI (parameters passed on command line CLI-style)
Progress
Slicer4 developer beta supports PythonQt scripted modules.
References
Example LabelStatistics module source code.
| . | . | . | . | . |
Command line to run "Slicelet" from Slicer4 build directory:
% Slicer4 lib/Slicer3/QTScriptedModules/LabelStatistics.py
- TODO Items:
- pass command line options to slicelets (use tclap-style --xml like the CLI modules)
- optimize startup time for the interpreter
- expose remaining parts of the Qt/qSlicer API to Python
- progress and status output
- wrapper to run non-scripted modules in slicelet mode
- Architectural Issues
- Object inheritance plan for scripted modules and slicelets
- Utilities needed to make it easy for developers/users to put together pipelines that mix module types