Difference between revisions of "2012 Summer Project Week:ANTS Registation"
From NAMIC Wiki
(Added ANTS webpage and image) |
(→References: Added Insight Journal reference) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
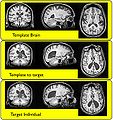
Image:ANTSLargeDef.jpg|ANTS Brain registration to population-based template | Image:ANTSLargeDef.jpg|ANTS Brain registration to population-based template | ||
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * Iowa: Dave Welch, Hans Johnson |
| − | * | + | * Penn State: Brian Avants, Gang Song, Nick Tustison |
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 20: | Line 13: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | We are developing | + | We are developing an ANTS-based registration module. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| Line 33: | Line 21: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | Our approach for | + | Our approach for implementing ANTS registration will be to create a command line module and integrate ANTS into the Slicer build process, if possible. |
| − | |||
| − | |||
</div> | </div> | ||
| Line 42: | Line 28: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | |||
| − | |||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
| − | |||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
| Line 54: | Line 36: | ||
This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below) | This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below) | ||
| − | #ITK Module | + | #ITK Module |
#Slicer Module | #Slicer Module | ||
##Built-in | ##Built-in | ||
| − | ##Extension -- commandline | + | ##Extension -- commandline YES |
##Extension -- loadable | ##Extension -- loadable | ||
#Other (Please specify) | #Other (Please specify) | ||
==References== | ==References== | ||
| − | * | + | * ANTS webpage: http://www.picsl.upenn.edu/ANTS/ [http://www.picsl.upenn.edu/ANTS/] |
| − | * | + | * Avants B, Tustison N, and Song G, ANTS: Open-Source Tools for Normalization And Neuroanatomy, IEEE Transactions on Medical Imaging, pp. 1-11. |
| − | * | + | * Avants B B, Tustison N T, Song G, Cook P A, Klein A, and Gee J C, A reproducible evaluation of ANTs similarity metric performance in brain image registration., NeuroImage, vol. 54, no. 3, pp. 2033-44, Feb. 2011. |
| − | + | * Avants B, Tustison N, and Song G, Advanced Normalization Tools: v1.0, Insight Journal, July 2009 [http://hdl.handle.net/10380/3113] | |
| − | * | ||
| − | |||
| − | |||
Latest revision as of 20:40, 22 May 2012
Home < 2012 Summer Project Week:ANTS RegistationKey Investigators
- Iowa: Dave Welch, Hans Johnson
- Penn State: Brian Avants, Gang Song, Nick Tustison
Objective
We are developing an ANTS-based registration module.
Approach, Plan
Our approach for implementing ANTS registration will be to create a command line module and integrate ANTS into the Slicer build process, if possible.
Progress
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below)
- ITK Module
- Slicer Module
- Built-in
- Extension -- commandline YES
- Extension -- loadable
- Other (Please specify)
References
- ANTS webpage: http://www.picsl.upenn.edu/ANTS/ [1]
- Avants B, Tustison N, and Song G, ANTS: Open-Source Tools for Normalization And Neuroanatomy, IEEE Transactions on Medical Imaging, pp. 1-11.
- Avants B B, Tustison N T, Song G, Cook P A, Klein A, and Gee J C, A reproducible evaluation of ANTs similarity metric performance in brain image registration., NeuroImage, vol. 54, no. 3, pp. 2033-44, Feb. 2011.
- Avants B, Tustison N, and Song G, Advanced Normalization Tools: v1.0, Insight Journal, July 2009 [2]