Difference between revisions of "2012 Summer Project Week:DifficultRegistration"
(Created page with '__NOTOC__ <gallery> Image:PW-MIT2012.png|Projects List Image:genuFAp.jpg|Scatter plot of the original FA data through the genu of the corpus…') |
m (Text replacement - "[http://www.na-mic.org/Wiki/images/" to "[https://na-mic.org/w/images/") |
||
| (62 intermediate revisions by 7 users not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2012.png|[[2012_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2012.png|[[2012_Summer_Project_Week#Projects|Projects List]] | ||
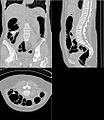
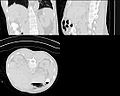
| − | Image: | + | Image:Ct-body-atlas.jpg |
| − | Image: | + | Image:Ct-body-cropped.jpg |
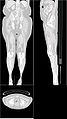
| + | Image:Ct-body-legs.jpg | ||
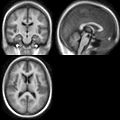
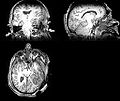
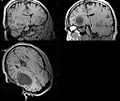
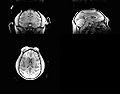
| + | Image:Mr-brain-atlas.jpg | ||
| + | Image:Mr-brain-tbi.jpg | ||
| + | Image:Mr-brain-rotated.jpg | ||
| + | Image:Mr-brain-rhesus.jpg | ||
</gallery> | </gallery> | ||
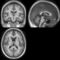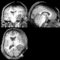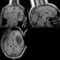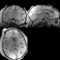
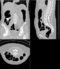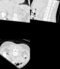
| − | == | + | ==Registration Result Visualization== |
| − | + | ||
| − | + | <b>Slicer:BRAINS<br>[[Image:IntraOp_Slicer-BRAINS_BSpline.gif|400px|lleft|IntraOp via BRAINS]]<br> | |
| − | + | NiftyReg<br>[[Image:mr-results-nifityreg.gif|400px|lleft|IntraOp via BRAINS]]<br> | |
| − | + | SIFT Landmark<br>[[Image:mr-results-sift.gif|200px|lleft|IntraOp via BRAINS]] [[Image:ct-results-sift.gif|200px|lleft|IntraOp via BRAINS]]</b><br> | |
| − | |||
==Key Investigators== | ==Key Investigators== | ||
| − | * UNC: | + | * Erasmus Medical Center: Stefan Klein |
| − | * | + | * University College London: Marc Modat |
| + | * UNC: Aditya Gupta, Martin Styner | ||
| + | * BWH: Matthew Toews, Petter Risholm, Dominik Meier, William Wells | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 21: | Line 27: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | To identify solutions to difficult image registration problems that challenge the limits of current technology. Aspects of difficulty will include: | |
| − | + | <ul> | |
| − | + | <li>inter-subject registration | |
| − | + | <li>truncation, missing tissue | |
| − | + | <li>unknown initialization | |
| − | + | <li>inter-species registration | |
| − | + | <li>articulated deformation | |
| + | </ul> | ||
</div> | </div> | ||
| Line 33: | Line 40: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | A set of difficult pair-wise registration problems will be considered. Participants will discuss workable solutions based on their expertise and background, and these solutions will be documented. | |
| − | + | <br> | |
| − | + | Registration cases can be found [http://www.matthewtoews.com/namic2012 here]. | |
| − | + | <br> | |
| − | + | *CT: atlas and torso have flipped orientation: Correction Xforms here: [[Media:FlipCT_Atlas+Torso.zip|Flip CT atlas&torso, zip file with 2 .tfm files]] | |
</div> | </div> | ||
| Line 43: | Line 50: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | We held a general discussion of registration challenges in the community. Results were generated for head and neck data (Ivan Kolesov), three registration approaches were evaluated on challenging head and body images: (NiftyReg - Marc Modat), (Slicer:BRAINS - Dominik Meier), (SIFT Landmarks - Matthew Toews). | |
| − | + | <ul> | |
| − | + | <li>Registration of difficult brain images is successful, body images remain challenging. | |
| + | <li>Registration direction has an important impact, i.e. which images are designed as fixed or moving. Symmetric or bi-directional registration is helpful. | ||
| + | <li>Masking of problematic image regions, e.g. pathology, is an important but time-consuming pre-processing task that could be further investigated or automated. For instance, interactive masking/segmentation of pathology, linking registration and masking. | ||
| + | </ul> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 52: | Line 62: | ||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
| + | <b>Attention Participants:</b> Please log in and update/correct entries in the table below. For bonus points, please provide links or solutions to image registration problems on this page.<br> | ||
| + | <table border="1"> | ||
| + | <th>Participant</th><th>Affiliation</th><th>Context</th><th>Techniques</th><th>Solutions/Links | ||
| + | </th><tr><td> | ||
| + | Steven Aylward</td><td>Kitware</td><td>CT, US, ressection, brain tumors, changing pathology, sliding organ</td><td>Sliding Geometry, Geometric Metamorphosis</td><td> | ||
| + | </td><tr><td> | ||
| + | Karl Diedrich</td><td>AZE</td><td>Rigid registration, abdomen</td><td>Multi-resolution registration</td><td> | ||
| + | </td><tr><td> | ||
| + | James Fishbaugh</td><td>SCI</td><td>Shape analysis and registration</td><td></td><td> | ||
| + | </td><tr><td> | ||
| + | Aditya Gupta</td><td>UNC</td><td>DTI, enlarged lateral ventricles</td><td></td><td> | ||
| + | </td><tr><td> | ||
| + | Stefan Klein</td><td>Erasmus Medical Center</td><td>General Registration</td><td></td><td> | ||
| + | </td><tr><td> | ||
| + | Ivan Kolesov</td><td>Georgia Tech</td><td>Articulated, point-based registration</td><td> Plastimatch, NiftyReg </td><td> [http://www.na-mic.org/Wiki/index.php/2012_Summer_Project_Week:Deformable_Registration_for_Head_and_Neck Head Neck Results] | ||
| + | </td><tr><td> | ||
| + | Dominik Meier</td><td>BWH</td><td>General registration</td><td>Slicer: BRAINS</td><td> | ||
| + | IntraOp:[[Image:IntraOp_Slicer-BRAINS_BSpline.gif|60px|lleft|IntraOp via BRAINS]]<br> | ||
| + | TBI:[[Image:TBI_Slicer-BRAINS_BSpline.gif|60px|lleft|TBI via BRAINS]]<br> | ||
| + | CT torso: FAILED</td><tr><td> | ||
| + | Marc Modat</td><td>UCL</td><td>neuro-deg. diseases, longitudinal</td><td>NiftiReg</td><td>[https://na-mic.org/w/images/d/d5/NiftyReg_projectWeek_test.pdf Some test res]</td><tr><td> | ||
| + | Albert Motillo</td><td>GE</td><td>Parsing CT, Detection</td><td></td><td> | ||
| + | </td><tr><td> | ||
| + | Simrin Nagpal</td><td>Queens University</td><td>CT/US registration</td><td></td><td> | ||
| + | </td><tr><td> | ||
| + | Samon Nuranian</td><td>UBC</td><td>US-guided intervension, spine</td><td></td><td> | ||
| + | </td><tr><td> | ||
| + | Andre Remi</td><td>UCLA</td><td>Longitudinal changes in TBI, tissue types</td><td></td><td> | ||
| + | </td><tr><td> | ||
| + | Peter Risholm</td><td>BWH</td><td>Brain, head and neck, radiation therapy</td><td>Probabilistic Uncertainty</td><td> | ||
| + | </td><tr><td> | ||
| + | Samira Sojoudi</td><td>UBC</td><td>Spine, CT/US registration</td><td></td><td> | ||
| + | </td><tr><td> | ||
| + | Matthew Toews</td><td>BWH</td><td>General registration</td><td>SIFT landmark correspondence</td><td> | ||
| + | MR-brain:[[Image:mr-results-sift.gif|60px|lleft]]<br> | ||
| + | CT-torso:[[Image:ct-results-sift.gif|60px|lleft]]<br> | ||
| + | </td><tr><td> | ||
| + | Bo Wang</td><td>Utah</td><td>TBI image segmentation</td><td></td><td> | ||
| + | </td><tr><td> | ||
| + | Kevin Wang</td><td>Princess Margaret Hospital</td><td>Adaptive radiation therapy, longitudinal</td><td></td><td> | ||
| + | </td><tr><td> | ||
| + | William Wells</td><td>BWH</td><td>Interventional applications</td><td>Theory: Segmentation, Registration</td><td></td> | ||
| + | <table> | ||
| − | + | == Links for Data == | |
| + | * [http://www.matthewtoews.com/namic2012 Matt's initial examples] | ||
| + | * [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:RegLibTable Slicer Registration Case Library] | ||
| + | * [http://dmip1.rad.jhmi.edu/xcat/ XCAT] | ||
| + | * [http://wiki.na-mic.org/Wiki/index.php/File:EnlargedLVCase_Normal.zip DTI Files Enlarged LV registration with normal control] | ||
| + | * [http://wiki.na-mic.org/Wiki/index.php/File:Krabbe_Controls_DWI.zip DWI Files Enlarged LV registration with normal control] | ||
| + | * [http://www.na-mic.org/Wiki/index.php/DBP3:UCLA#Data TBI Cases] | ||
| + | * [http://www.nitrc.org/projects/tumorsim/ TumorSim] longitudinal data: To appear at http://midas3.kitware.com | ||
| − | + | == Links for Tools & Methods == | |
| − | + | * Sliding Geometries Registration: http://public.kitware.com/Wiki/TubeTK | |
| − | + | * Geometric Metamorphosis: https://github.com/calaTK/calaTK | |
| − | |||
| − | |||
| − | |||
| − | == | + | == Links for Papers == |
| − | * | + | * Sliding Geometries |
| − | * | + | ** ISBI 2011: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3141338/ |
| − | * | + | ** Abdominal Imaging, MICCAI, 2011: http://www.springerlink.com/content/552824638l375645/ |
| − | * | + | * Geometric MetaMorphosis |
| + | ** MICCAI 2011: http://www.springerlink.com/content/7r077665012078r5/ | ||
</div> | </div> | ||
Latest revision as of 18:27, 10 July 2017
Home < 2012 Summer Project Week:DifficultRegistrationRegistration Result Visualization
Slicer:BRAINS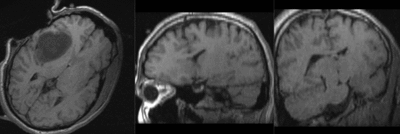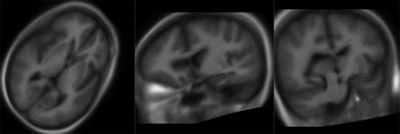
NiftyReg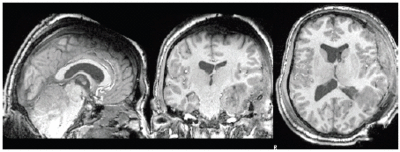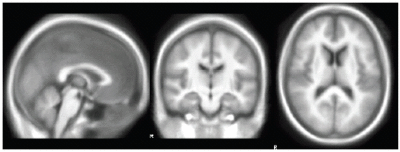
SIFT Landmark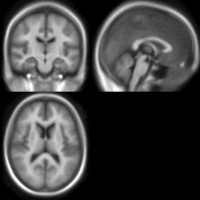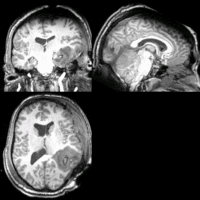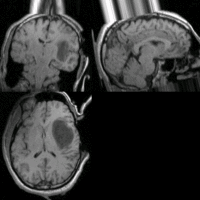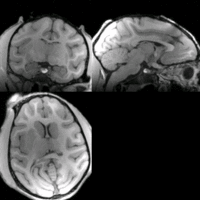
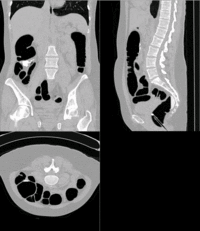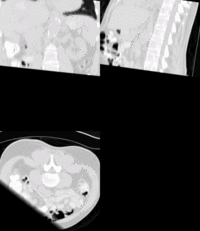
Key Investigators
- Erasmus Medical Center: Stefan Klein
- University College London: Marc Modat
- UNC: Aditya Gupta, Martin Styner
- BWH: Matthew Toews, Petter Risholm, Dominik Meier, William Wells
Objective
To identify solutions to difficult image registration problems that challenge the limits of current technology. Aspects of difficulty will include:
- inter-subject registration
- truncation, missing tissue
- unknown initialization
- inter-species registration
- articulated deformation
Approach, Plan
A set of difficult pair-wise registration problems will be considered. Participants will discuss workable solutions based on their expertise and background, and these solutions will be documented.
Registration cases can be found here.
- CT: atlas and torso have flipped orientation: Correction Xforms here: Flip CT atlas&torso, zip file with 2 .tfm files
Progress
We held a general discussion of registration challenges in the community. Results were generated for head and neck data (Ivan Kolesov), three registration approaches were evaluated on challenging head and body images: (NiftyReg - Marc Modat), (Slicer:BRAINS - Dominik Meier), (SIFT Landmarks - Matthew Toews).
- Registration of difficult brain images is successful, body images remain challenging.
- Registration direction has an important impact, i.e. which images are designed as fixed or moving. Symmetric or bi-directional registration is helpful.
- Masking of problematic image regions, e.g. pathology, is an important but time-consuming pre-processing task that could be further investigated or automated. For instance, interactive masking/segmentation of pathology, linking registration and masking.
Delivery Mechanism
Attention Participants: Please log in and update/correct entries in the table below. For bonus points, please provide links or solutions to image registration problems on this page.
| Participant | Affiliation | Context | Techniques | Solutions/Links |
|---|---|---|---|---|
| Steven Aylward | Kitware | CT, US, ressection, brain tumors, changing pathology, sliding organ | Sliding Geometry, Geometric Metamorphosis | |
| Karl Diedrich | AZE | Rigid registration, abdomen | Multi-resolution registration | |
| James Fishbaugh | SCI | Shape analysis and registration | ||
| Aditya Gupta | UNC | DTI, enlarged lateral ventricles | ||
| Stefan Klein | Erasmus Medical Center | General Registration | ||
| Ivan Kolesov | Georgia Tech | Articulated, point-based registration | Plastimatch, NiftyReg | Head Neck Results |
| Dominik Meier | BWH | General registration | Slicer: BRAINS | CT torso: FAILED |
| Marc Modat | UCL | neuro-deg. diseases, longitudinal | NiftiReg | Some test res |
| Albert Motillo | GE | Parsing CT, Detection | ||
| Simrin Nagpal | Queens University | CT/US registration | ||
| Samon Nuranian | UBC | US-guided intervension, spine | ||
| Andre Remi | UCLA | Longitudinal changes in TBI, tissue types | ||
| Peter Risholm | BWH | Brain, head and neck, radiation therapy | Probabilistic Uncertainty | |
| Samira Sojoudi | UBC | Spine, CT/US registration | ||
| Matthew Toews | BWH | General registration | SIFT landmark correspondence | |
| Bo Wang | Utah | TBI image segmentation | ||
| Kevin Wang | Princess Margaret Hospital | Adaptive radiation therapy, longitudinal | ||
| William Wells | BWH | Interventional applications | Theory: Segmentation, Registration |
Links for Data
- Matt's initial examples
- Slicer Registration Case Library
- XCAT
- DTI Files Enlarged LV registration with normal control
- DWI Files Enlarged LV registration with normal control
- TBI Cases
- TumorSim longitudinal data: To appear at http://midas3.kitware.com
Links for Tools & Methods
- Sliding Geometries Registration: http://public.kitware.com/Wiki/TubeTK
- Geometric Metamorphosis: https://github.com/calaTK/calaTK
Links for Papers
- Sliding Geometries
- ISBI 2011: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3141338/
- Abdominal Imaging, MICCAI, 2011: http://www.springerlink.com/content/552824638l375645/
- Geometric MetaMorphosis