Difference between revisions of "2016 Winter Project Week/Projects/DigitalPathologyNuclearSegmentation"
From NAMIC Wiki
| Line 38: | Line 38: | ||
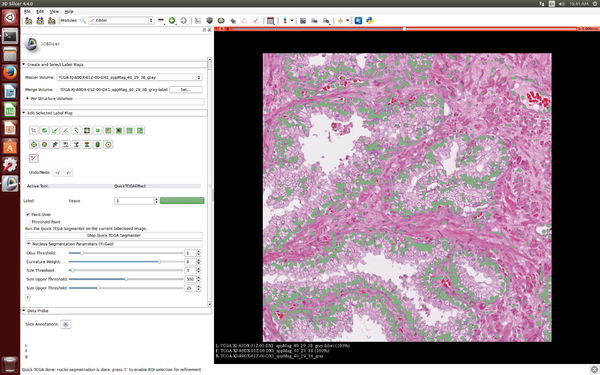
[[File:SlicerPathologybeginworkweek2016.png|600px|thumb|left|SlicerPathology extension at the start of Winter Work Week 2016]] | [[File:SlicerPathologybeginworkweek2016.png|600px|thumb|left|SlicerPathology extension at the start of Winter Work Week 2016]] | ||
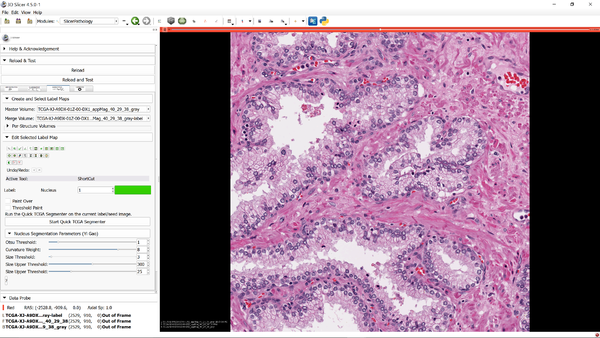
| + | [[File:Afterworkweek.png|600px|thumb|left|SlicerPathology extension at the end of Winter Work Week 2016]] | ||
==Background and References== | ==Background and References== | ||
Revision as of 04:49, 8 January 2016
Home < 2016 Winter Project Week < Projects < DigitalPathologyNuclearSegmentationKey Investigators
- Erich Bremer - Stony Brook University
- Yi Gao - Stony Brook University
- Nicole Aucoin - Brigham and Women's Hospital
- Andrey Fedorov - Brigham and Women's Hospital
- Kelli Xu - MIT
Project Description
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
|