Difference between revisions of "2016 Winter Project Week/Projects/DigitalPathologyNuclearSegmentation"
From NAMIC Wiki
| (20 intermediate revisions by 3 users not shown) | |||
| Line 7: | Line 7: | ||
==Key Investigators== | ==Key Investigators== | ||
<!-- Add a bulleted list of investigators and their institutions here --> | <!-- Add a bulleted list of investigators and their institutions here --> | ||
| + | |||
| + | * Erich Bremer - [https://bmi.stonybrookmedicine.edu/ Stony Brook University] | ||
| + | * Yi Gao - [https://bmi.stonybrookmedicine.edu/ Stony Brook University] | ||
| + | * Nicole Aucoin - [http://www.brighamandwomens.org/ Brigham and Women's Hospital] | ||
| + | * Andrey Fedorov - [http://www.brighamandwomens.org/ Brigham and Women's Hospital] | ||
==Project Description== | ==Project Description== | ||
{| class="wikitable" | {| class="wikitable" | ||
| + | ! style="text-align: left; width:27%" | Objective | ||
| + | ! style="text-align: left; width:27%" | Approach and Plan | ||
| + | ! style="text-align: left; width:27%" | Progress and Next Steps | ||
|- style="vertical-align:top;" | |- style="vertical-align:top;" | ||
| − | + | | | |
| − | <!-- | + | <!-- Objective bullet points --> |
| − | * | + | * Create intuitive and user-friendly GUI |
| − | + | * make meta-data exportable | |
| − | + | * Prepare extension for Phase II cloud work and future modules | |
| − | <!-- | + | | |
| − | * | + | <!-- Approach and Plan bullet points --> |
| − | + | * create work-flow module walking user through needed steps | |
| − | + | * wrap editor module to allow for customization for segmentation purposes without interfering with global editor instance | |
| − | <!-- | + | * capture needed data elements and export them as [http://www.w3.org/TR/json-ld/ JSON-LD] supplementing exporting of masks |
| − | * | + | * augment GUI by requesting authentication information for [http://oauth.net/2/ OAuth 2.0] and adding elements for image selection via web. |
| + | * refactor existing extension modules into separate distinct editor effects modules (currently lumped as one) | ||
| + | | | ||
| + | <!-- Progress and Next steps bullet points (fill out at the end of project week --> | ||
| + | * initial work-flow module created | ||
| + | * editor module wrapped and including in one of the step tabs of the work-flow module | ||
| + | * label mask saved, still need to add additional meta data (algorithm parameters) as JSON-LD export | ||
| + | * username/password entries added to work-flow module for future web authentication | ||
| + | * original single editor-effect refactored into separate effects. Further cleaning of of code needed | ||
| + | * future work - allow image tiles to be select via web interface and resultant label masks and meta data to be exported directly to web services using OAuth2 authenication | ||
| + | * remove unneeded GUI elements and set additional field automatically. Create better tab icons! | ||
| + | * test SlicerPathology with new OpenCV Extension | ||
| + | |} | ||
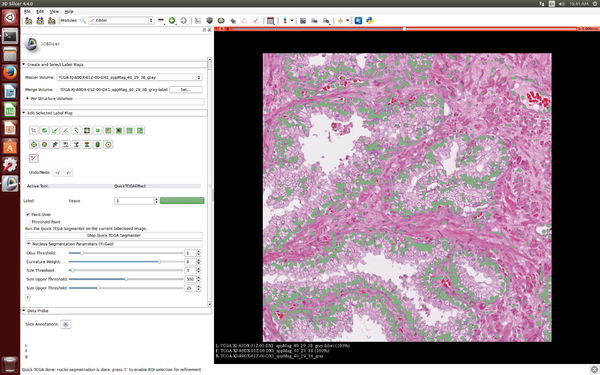
| − | | | + | [[File:SlicerPathologybeginworkweek2016.png|600px|thumb|left|SlicerPathology extension at the start of Winter Work Week 2016]] |
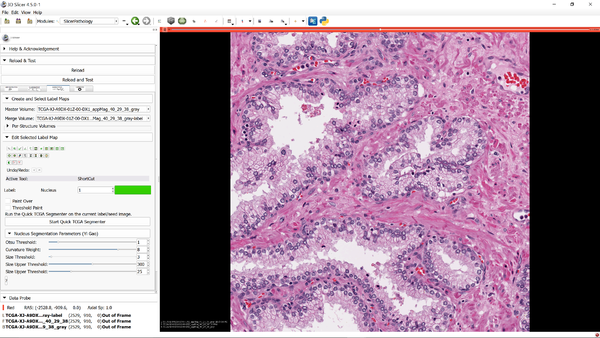
| + | [[File:Afterworkweek.png|600px|thumb|left|SlicerPathology extension at the end of Winter Work Week 2016]] | ||
==Background and References== | ==Background and References== | ||
<!-- Use this space for information that may help people better understand your project, like links to papers, source code, or data --> | <!-- Use this space for information that may help people better understand your project, like links to papers, source code, or data --> | ||
| + | |||
| + | * [https://github.com/SBU-BMI/SlicerPathology SlicerPathology extension source repository] | ||
| + | * [http://interactivemedical.org/imic2014/CameraReadyPapers/Paper%204/IMIC_ID4_FastGrowCut.pdf Fast Growcut] | ||
Latest revision as of 13:56, 8 January 2016
Home < 2016 Winter Project Week < Projects < DigitalPathologyNuclearSegmentationKey Investigators
- Erich Bremer - Stony Brook University
- Yi Gao - Stony Brook University
- Nicole Aucoin - Brigham and Women's Hospital
- Andrey Fedorov - Brigham and Women's Hospital
Project Description
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
|