2016 Winter Project Week/Projects/DigitalPathologyNuclearSegmentation
From NAMIC Wiki
Home < 2016 Winter Project Week < Projects < DigitalPathologyNuclearSegmentation
Key Investigators
- Erich Bremer - Stony Brook University
- Yi Gao - Stony Brook University
- Nicole Aucoin - Brigham and Women's Hospital
- Andrey Fedorov - Brigham and Women's Hospital
- Kelli Xu - MIT
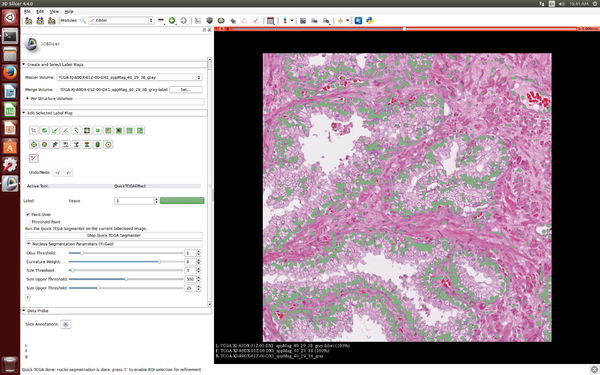
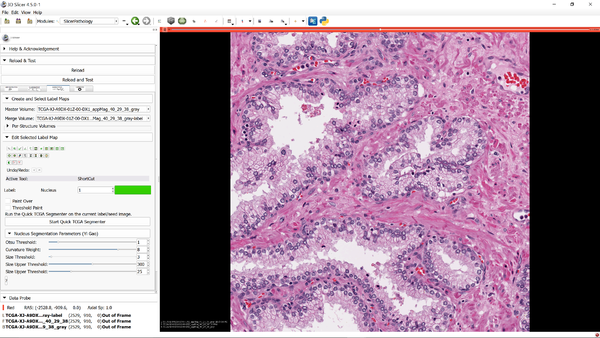
Project Description
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
|