AHM2009:ExternalWFU
External Collaboration: Measuring Alcohol and Stress Interactions with Structural and Perfusion MRI
Wake Forest Collaboration Update (ppt)
Introduction
Our goal is to use MRI to examine the effects of chronic alcohol self-administration on brain structure and function in a group of monkeys that have been mother-reared or nursery-reared.
Team
- Jim Daunais (WFUHS PI)
- Bob Kraft (WFUHS MR Physicist)
- Chris Wyatt (VT Engineering)
- Vidya Rajagopalan (VT Engineering)
- Killian Pohl (NA-MIC Core 1)
Goals for 2008
- Acquisition of baseline vervet images for atlas development (completed)
- Vervet atlas completed (completed)
- Animal Training and Induction 3-4 months (completed)
Methods
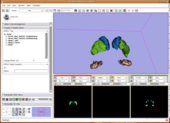
The first stage of atlas construction was based on a method by Styner et al [1]. A template image was chosen randomly from the training images. This image was manually skull-stripped and a skull-strip mask determined. All other training images were histogram matched and then affinely registered to this template using FSL FLIRT [2] and skull-stripped using the skull-strip mask after dilation by one voxel. The images of only the Extra Cranial Content (ECC) are averaged to form the ECC template.
The Intra Cranial Content (ICC) images of the various subjects are again affinely registered to the ICC image of the chosen template using FSL FLIRT [2]. The affinely registered images were deformably registered to the same target using the diffeomorphic demons registration method. The registered images were then averaged. The ICC average was now combined with the ECC template to form the template image for the whole head.
This high-CNR template image was then segmented into gray matter, white matter, and CSF tissue classes using initial k-means clustering and manual correction. Subcortical structures were then manually traced on the high-CNR template. The tissue and structure segmentations were smoothed using a Gaussian kernel to obtain the initial spatial probability maps.
The second stage of the construction used the initial spatial probability maps from stage 1 with the expectation-maximization segmentation (EMSegmenter) method by Pohl et al. [3]. Using the initial spatial probabilities the EMS algorithm, the initial spatial probability maps were combined with manually estimated structure intensity distributions for each structure to produce the final segmentation for the template image. This segmentation was then manually corrected for errors and then was smoothed using a guassian kernel of variance 1.
[1] M. Styner, R. Knickmeyer, S. Joshi, C. Coe, S. J. Short, and J. Gilmore. Automatic brain segmentation in rhesus monkeys. Proc SPIE Medical Imaging Conference, Proc SPIE Vol 6512 Medical Imaging 2007, pp 65122L-1 - 65122L-8.
[2] M. Jenkinson and S.M. Smith. A global optimisation method for robust affine registration of brain images. Medical Image Analysis, 5(2):143-156, June 2001.
[3] Pohl KM, Fisher J, Levitt JJ, Shenton ME, Kikinis R, Grimson WEL, Wells WM. A unifying approach to registration, segmentation, and intensity correction. Medical Image Computing And Computer-Assisted Intervention - MICCAI 2005; PT 1 Lecture Notes In Computer Science 3749: 310-318.
Products
Future Work
- Animal ad-lib access to alcohol for 12 months
- Final structural scans ~August 2009