Difference between revisions of "AHM2009:JHU"
Sidd queens (talk | contribs) |
Sidd queens (talk | contribs) |
||
| Line 13: | Line 13: | ||
* The steps in the workflow are as follows: | * The steps in the workflow are as follows: | ||
** Calibration, to register the image to the biopsy device via MR fiducials. | ** Calibration, to register the image to the biopsy device via MR fiducials. | ||
| + | ** Segmentation, to segment the prostate from the MRI scan (Algorithm by Yi Gao/ Allen Tannenbaum GeorgiaTech) | ||
** Targeting, to mark biopsy targets (manually) and compute needle trajectory and depth (automatically). | ** Targeting, to mark biopsy targets (manually) and compute needle trajectory and depth (automatically). | ||
** Verification, to use post-biopsy images to verify biopsy locations against the planned targets. | ** Verification, to use post-biopsy images to verify biopsy locations against the planned targets. | ||
| Line 18: | Line 19: | ||
==Detailed Information about the Pipeline== | ==Detailed Information about the Pipeline== | ||
| − | + | ;* What problem does the pipeline solve? | |
There are several Slicer features that are crucial to image-guided therapy that are utilized in this module: | There are several Slicer features that are crucial to image-guided therapy that are utilized in this module: | ||
| Line 32: | Line 33: | ||
The module uses optical encoders that are attached to the joints of the biopsy device to verify that the position of the device matches that of the plan. | The module uses optical encoders that are attached to the joints of the biopsy device to verify that the position of the device matches that of the plan. | ||
| + | |||
| + | ;* Who is the targeted user? | ||
| + | |||
| + | ;* How does the pipeline compare to state of the art? | ||
==Software & documentation== | ==Software & documentation== | ||
Revision as of 15:24, 7 January 2009
Home < AHM2009:JHU
JHU Roadmap Project
Overview
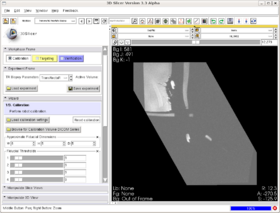
We have created a module that provides a workflow interface for MR-guided transrectal prostate biopsy. The MR images are captured with the help of an endorectal coil which is mounted on the same shaft as the biopsy needle.
- The steps in the workflow are as follows:
- Calibration, to register the image to the biopsy device via MR fiducials.
- Segmentation, to segment the prostate from the MRI scan (Algorithm by Yi Gao/ Allen Tannenbaum GeorgiaTech)
- Targeting, to mark biopsy targets (manually) and compute needle trajectory and depth (automatically).
- Verification, to use post-biopsy images to verify biopsy locations against the planned targets.
This module provides a demonstration of how Slicer modules can be created for specific interventional devices.
Detailed Information about the Pipeline
- What problem does the pipeline solve?
There are several Slicer features that are crucial to image-guided therapy that are utilized in this module:
Oriented volumes and image slice reformatting
Each volume acquired during the biopsy procedure has its own orientation, since images are acquired according to the orientation of the instrument, which is at an oblique angle to the MR scanner's coordinate axes. What we have added is that, for each workflow step, a particular volume is specified as the "primary" and, when overlays are performed e.g. for verification, Slicer displays the primary image in its original orientation and reslices the others. The displayed slice orientation automatically changes to match the primary whenever the workflow step changes.
Multiple fiducial lists
This module maintains two Slicer fiducial lists: for registration, and for targeting. Like the image orientation, we have added automatic switching between display of fiducials according to the workflow step.
Communication with hardware devices
The module uses optical encoders that are attached to the joints of the biopsy device to verify that the position of the device matches that of the plan.
- Who is the targeted user?
- How does the pipeline compare to state of the art?
Software & documentation
- The TRProstateBiopsy module is in the "Queens" directory of the NAMICSandBox - access online
- Tutorial is forthcoming
Team
- DBP: Queens School of Computing, Queens University ; LCSR, Johns Hopkins University
- Core 1: Affiliation & logo
- Core 2: Affiliation & logo
- Contact: name, email
Outreach
- Publication Links to the PubDB.
- Planned outreach activities (including presentations, tutorials/workshops) at conferences