AHM2009:JHU
JHU Roadmap Project
Overview
- Who is the targeted user?
The only definitive method to diagnose prostate cancer is to perform biopsy. The current gold standard is Trans Rectal UltraSound (TRUS) guided biopsies. TRUS biopsies lack in sensitivity and specificity. MRI has recently been investigated as an attractive alternative to image and localize prostate cancer. It is imperative to take advantage of multi-parametric MRI imaging to perform prostate biopsy. However, to perform MRI-guided biopsy, there is a physical limitation of working in a very small space. We have developed a completely MR-compatible robot that solves the problem. This SLICER based module we are developing, aims to provide end-to-end interventional solution that combines software imaging functionality and interfaces with our specific hardware to perform the biopsy. The targeted users are the clinicians who are currently investigating our MRI compatible robotic device.
- What problem does the pipeline solve?
There are several Slicer features that are crucial to image-guided therapy that are utilized in this module:
Oriented volumes and image slice reformatting Each volume acquired during the biopsy procedure has its own orientation, since images are acquired according to the orientation of the instrument, which is at an oblique angle to the MR scanner's coordinate axes. What we have added is that, for each workflow step, a particular volume is specified as the "primary" and, when overlays are performed e.g. for verification, Slicer displays the primary image in its original orientation and reslices the others. The displayed slice orientation automatically changes to match the primary whenever the workflow step changes.
Multiple fiducial lists This module maintains two Slicer fiducial lists: for registration, and for targeting. Like the image orientation, we have added automatic switching between display of fiducials according to the workflow step.
Communication with hardware devices The module uses optical encoders that are attached to the joints of the biopsy device to verify that the position of the device matches that of the plan.
- How does the pipeline compare to state of the art?
SLICER's ability to work with volumes, slice reformatting, and seamless integration with image analysis algorithms makes it uniquely suited to our problem. One does not have to reinvent the wheel, as there is so much functionality that is available at hand and easily accessible as plug and play components. There is an existing pipeline/application that interfaces with our specific hardware, however, it is rather difficult to extend its functionality to cater near future requirements. So, we believe SLICER module is the way to go for us.
Detailed Information about the Pipeline
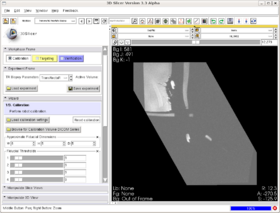
We have created a module that provides a workflow interface for MR-guided transrectal prostate biopsy. The MR images are captured with the help of an endorectal coil which is mounted on the same shaft as the biopsy needle.
- The steps in the workflow are as follows:
- Calibration, to register the image to the biopsy device via MR fiducials.
- Segmentation, to segment the prostate from the MRI scan (Algorithm by Yi Gao/ Allen Tannenbaum GeorgiaTech)
- Targeting, to mark biopsy targets (manually) and compute needle trajectory and depth (automatically).
- Verification, to use post-biopsy images to verify biopsy locations against the planned targets.
This module provides a demonstration of how Slicer modules can be created for specific interventional devices.
Software & documentation
- The TRProstateBiopsy module is in the "Queens" directory of the NAMICSandBox - access online
- Tutorial is forthcoming
Team
- DBP: Queens School of Computing, Queens University ; LCSR, Johns Hopkins University
- Core 1: Affiliation & logo
- Core 2: Affiliation & logo
- Contact: name, email
Outreach
- Publication Links to the PubDB.
- Planned outreach activities (including presentations, tutorials/workshops) at conferences