Difference between revisions of "AHM2009:UNC"
| Line 5: | Line 5: | ||
{| | {| | ||
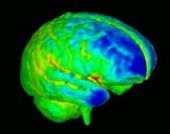
| − | |[[Image: | + | |[[Image:BrainDevelopment.jpg|thumb|170px|Brain development]] |
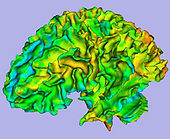
| − | |[[Image: | + | |[[Image:WMThickness.jpg|thumb|170px|Cortical thickness on WM surface]] |
| − | |||
|} | |} | ||
== Overview == | == Overview == | ||
* What problem does the pipeline solve, and who is the targeted user? | * What problem does the pipeline solve, and who is the targeted user? | ||
| + | |||
| + | There is a lack of appropriate tools for processing of pediatric MRI, a challenging topic since pediatric MRI differs significantly from adult MRI due to variable brain shape and the process of maturation/myelination which are reflected in nonlinear shape/volume changes but also regional change of white matter.... | ||
| + | |||
| + | We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional and local cortical thickness. Such a workflow applied to the young brain (2-4 years old) is one goal of the UNC DBP | ||
| + | |||
* How does the pipeline compare to state of the art | * How does the pipeline compare to state of the art | ||
| + | |||
| + | A statistical analysis is currently performed in order to compare our pipeline with FreeSurfer. We use a pediatric dataset of 90 cases including controls autistic children and developmental-delays, 2 groups being considered: 2 year-old cases and 4-year old cases. | ||
==Detailed Information about the Pipeline== | ==Detailed Information about the Pipeline== | ||
* Illustrate the components and workflow of the pipeline using your own data | * Illustrate the components and workflow of the pipeline using your own data | ||
* Demonstrate parameters/steps that need to be adjusted using someone else's data | * Demonstrate parameters/steps that need to be adjusted using someone else's data | ||
| + | |||
| + | |||
| + | * [[DBP2:UNC:Regional_Cortical_Thickness_Pipeline | Regional cortical thickness pipeline]] | ||
| + | |||
| + | * [[DBP2:UNC:Local_Cortical_Thickness_Pipeline | Local cortical thickness pipeline]] | ||
==Software & documentation== | ==Software & documentation== | ||
| − | * | + | * Executables and tutorial dataset: http://www.nitrc.org/projects/arctic/ |
| − | |||
==Team== | ==Team== | ||
| − | * DBP: | + | * DBP: University of North Carolina at Chapel Hill |
| − | * Core 1: | + | ** Co-PI: Heather Cody Hazlett, PhD |
| − | * Core 2: | + | ** Co-PI: Joseph Piven, MD |
| − | * Contact: | + | UNC DBP: Clement Vachet |
| + | * Core 1: Martin Styner, UNC Chapel Hill | ||
| + | ** UNC Algorithm: Ipek Oguz, Nicolas Augier, Marcel Prastawa, Marc Niethammer, Cedric Mathieu, Clement Vachet | ||
| + | * Core 2: Jim Miller, GE Research | ||
| + | * Contact: Heather Cody Hazlett, PhD, (heather_cody at med.unc.edu, Ph: 919-966-4099) | ||
==Outreach== | ==Outreach== | ||
* Publication Links to the PubDB. | * Publication Links to the PubDB. | ||
* Planned outreach activities (including presentations, tutorials/workshops) at conferences | * Planned outreach activities (including presentations, tutorials/workshops) at conferences | ||
Revision as of 01:24, 14 December 2008
Home < AHM2009:UNC
UNC Roadmap Project
Overview
- What problem does the pipeline solve, and who is the targeted user?
There is a lack of appropriate tools for processing of pediatric MRI, a challenging topic since pediatric MRI differs significantly from adult MRI due to variable brain shape and the process of maturation/myelination which are reflected in nonlinear shape/volume changes but also regional change of white matter....
We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional and local cortical thickness. Such a workflow applied to the young brain (2-4 years old) is one goal of the UNC DBP
- How does the pipeline compare to state of the art
A statistical analysis is currently performed in order to compare our pipeline with FreeSurfer. We use a pediatric dataset of 90 cases including controls autistic children and developmental-delays, 2 groups being considered: 2 year-old cases and 4-year old cases.
Detailed Information about the Pipeline
- Illustrate the components and workflow of the pipeline using your own data
- Demonstrate parameters/steps that need to be adjusted using someone else's data
Software & documentation
- Executables and tutorial dataset: http://www.nitrc.org/projects/arctic/
Team
- DBP: University of North Carolina at Chapel Hill
- Co-PI: Heather Cody Hazlett, PhD
- Co-PI: Joseph Piven, MD
UNC DBP: Clement Vachet
- Core 1: Martin Styner, UNC Chapel Hill
- UNC Algorithm: Ipek Oguz, Nicolas Augier, Marcel Prastawa, Marc Niethammer, Cedric Mathieu, Clement Vachet
- Core 2: Jim Miller, GE Research
- Contact: Heather Cody Hazlett, PhD, (heather_cody at med.unc.edu, Ph: 919-966-4099)
Outreach
- Publication Links to the PubDB.
- Planned outreach activities (including presentations, tutorials/workshops) at conferences