Algorithm:Utah
Back to NA-MIC Algorithms
Overview of Utah Algorithms (PI: Ross Whitaker)
We are developing new methods in the areas of statistical shape analysis, MRI tissue segmentation, and diffusion tensor image processing and analysis. We are building shape analysis tools that can generate efficient statistical models appropriate for analyzing anatomical shape differences in the brain. We are developing a wide range of tools for diffusion tensor imaging, that span the entire pipeline from image processing to automatic white matter tract extraction to statistical testing of clinical hypotheses.
Utah Projects

|
Particle Based Shape RegressionShape regression promises to be an important tool to study the relationship between anatomy and underlying clinical or biological parameters, such as age. We propose a new method to building shape models that incorporates regression analysis in the process of optimizing correspondences on a set of open surfaces. The method is applied to provide new results on clinical MRI data related to early development of the human head. New: M Datar, J Cates, P T Fletcher, S Gouttard, G Gerig, R Whitaker, Particle Based Shape Regression of Open Surfaces with Applications to Developmental Neuroimaging, MICCAI 2009. |
|
Brain Manifold LearningThis work is concerned with modeling the space of brain images. Common approach for representing populations are template or clustering based approaches. In this project we develop a data driven method to learn a manifold representation from a set of brain images. The presented approach is described and evaluated in the setting of brain MRI but generalizes to other application domains. New: S Gerber, T Tasdizen, S Joshi, R Whitaker, On the Manifold Structure of the Space of Brain Images, MICCAI 2009. New: S Gerber, T Tasdizen, R Whitaker, Dimensionality Reduction and Principal Surfaces via Kernel Map, ICCV 2009. |

|
Diffusion Tensor Image Processing ToolsWe implement the diffusion weighted image (DWI) registration model from the paper of G.K.Rohde et al. Patient head motion and eddy currents distortion cause artifacts in maps of diffusion parameters computer from DWI. This model corrects these two distortions at the same time including brightness correction. |
|
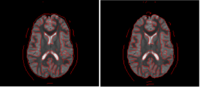
Correction for Geometric Distortion in Echo Planar ImagesWe have developed a variational image-based approach to correct the susceptibility artifacts in the alignment of diffusion weighted and structural MRI.The correction is formulated as an optimization of a penalty that captures the intensity difference between the jacobian corrected EPI baseline images and a corresponding T2-weighted structural image. New: R Tao, P T Fletcher, S Gerber, R Whitaker, A Variational Image-Based Approach to the Correction of Susceptibility Artifacts in the Alignment of Diffusion Weighted and Structural MRI, IPMI 2009. |
|
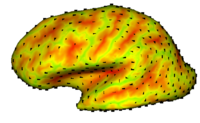
Cortical Correspondence using Particle SystemIn this project, we want to compute cortical correspondence on populations, using various features such as cortical structure, DTI connectivity, vascular structure, and functional data (fMRI). This presents a challenge because of the highly convoluted surface of the cortex, as well as because of the different properties of the data features we want to incorporate together. More... New: Oguz I, Niethammer M, Cates J, Whitaker R, Fletcher T, Vachet C, Styner M. “Cortical Correspondence with Probabilistic Fiber Connectivity”. Proc. Information Processing in Medical Imaging, 2009. |
|
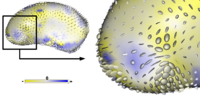
Adaptive, Particle-Based Sampling for Shapes and ComplexesThis research is a new method for constructing compact statistical point-based models of ensembles of similar shapes that does not rely on any specific surface parameterization. The method requires very little preprocessing or parameter tuning, and is applicable to a wider range of problems than existing methods, including nonmanifold surfaces and objects of arbitrary topology. More... New: Particle-Based Shape Analysis of Multi-object Complexes. Cates J., Fletcher P.T., Styner M., Hazlett H.C., Whitaker R. Int Conf Med Image Comput Comput Assist Interv. 2008;11(Pt 1):477-485. |

|
Shape Analysis Framework using SPHARM-PDMThe UNC shape analysis is based on an analysis framework of objects with spherical topology, described mainly by sampled spherical harmonics SPHARM-PDM. The input of the shape analysis framework is a set of binary segmentations of a single brain structure, such as the hippocampus or caudate. These segmentations are converted into a shape description (SPHARM) with correspondence and analyzed via Hotelling T^2 two sample metric. More... New: Zhao Z., Taylor W., Styner M., Steffens D., Krishnan R., Macfall J. , Hippocampus shape analysis and late-life depression. PLoS ONE. 2008 Mar 19;3(3):e1837.
|
|
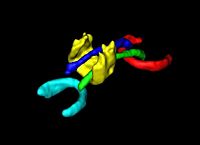
DTI Volumetric White Matter ConnectivityWe have developed a PDE-based approach to white matter connectivity from DTI that is founded on the principal of minimal paths through the tensor volume. Our method computes a volumetric representation of a white matter tract given two endpoint regions. We have also developed statistical methods for quantifying the full tensor data along these pathways, which should be useful in clinical studies using DT-MRI. More... |
|
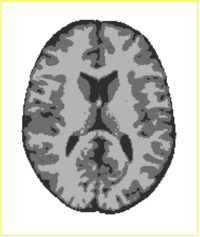
Tissue Classification with Neighborhood StatisticsWe have implemented an MRI tissue classification algorithm based on unsupervised non-parametric density estimation of tissue intensity classes. More... |