Difference between revisions of "Algorithm:Utah2"
| Line 8: | Line 8: | ||
{| cellpadding="10" style="text-align:left;" | {| cellpadding="10" style="text-align:left;" | ||
| + | |||
| + | | style="width:15%" | [[Image:TBIseg.png|200px]] | ||
| + | | style="width:85%" | | ||
| + | |||
| + | == [[Projects:PathologyAnalysis|Analysis of Brain Images with Pathological Changes ]] == | ||
| + | |||
| + | Quantification, analysis and display of brain pathology as observed in MRI is important for diagnosis, monitoring of disease progression, improved understanding of pathological processes and for studying treatment strategies. | ||
| + | We conduct research on developing novel methodologies that covers registration, segmentation, visualization of pathological structures | ||
| + | with the aim of quantifying changes due to lesions, bleeding, and deformations over time | ||
| + | |||
| + | |- | ||
| style="width:15%" | [[Image:Cbg-dtiatlas-tracts.png|200px]] | | style="width:15%" | [[Image:Cbg-dtiatlas-tracts.png|200px]] | ||
Revision as of 19:45, 18 March 2011
Home < Algorithm:Utah2Back to NA-MIC Algorithms
Overview of Utah 2 Algorithms (PI: Guido Gerig)
At Utah, we are interested in a range of algorithms and solutions for the analysis of DTI and the segmentation of healthy brains and brains with lupus lesions from structural MRI.
Utah 2 Projects
| 200px |
Analysis of Brain Images with Pathological ChangesQuantification, analysis and display of brain pathology as observed in MRI is important for diagnosis, monitoring of disease progression, improved understanding of pathological processes and for studying treatment strategies. We conduct research on developing novel methodologies that covers registration, segmentation, visualization of pathological structures with the aim of quantifying changes due to lesions, bleeding, and deformations over time |
|
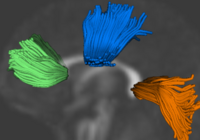
Group Analysis of DTI Fiber TractsAnalysis of populations of diffusion images typically requires time-consuming manual segmentation of structures of interest to obtain correspondance for statistics. This project uses non-rigid registration of DTI images to produce a common coordinate system for hypothesis testing of diffusion properties. More... New: Casey B. Goodlett, P. Thomas Fletcher, John H. Gilmore, Guido Gerig. Group Analysis of DTI Fiber Tract Statistics with Application to Neurodevelopment. NeuroImage 45 (1) Supp. 1, 2009. p. S133-S142. |
|
Lesion SegmentationQuantification, analysis and display of brain pathology such as white matter lesions as observed in MRI is important for diagnosis, monitoring of disease progression, improved understanding of pathological processes and for developing new therapies. More... New: Marcel Prastawa and Guido Gerig. Automatic MS Lesion Segmentation by Outlier Detection and Information Theoretic Region Partitioning. 3D Segmentation in the Clinic: A Grand Challenge II Workshop at Medical Image Computing and Computer Assisted Intervention (MICCAI) 2008. |

|
DTI fiber tract statisticsOngoing: As part of this project, we are processing DTI data and computing statistics along white matter fiber tracts. For this, we are building a command line tool which allows the user to study the behavior of water diffusion along the length of the tracts. More... |
|
Influence of Imaging Noise on DTI StatisticsClinical acquisition of diffusion weighted images with high signal to noise ratio remains a challenge. The goal of this project is to understand the impact of MR noise on quantiative statistics of diffusion properties such as anisotropy measures, trace, etc. More... |

|
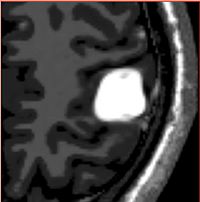
Atlas Based Brain SegmentationAutomatic segmentation can be performed reliably using priors from brain atlases and an image generative model. We have developed a tool that provides an automatic segmentation pipeline in a modular framework. More... |
|
Tumor Simulation for Validating Change Tracking ApplicationsDetermining extent of pathology as it changes over time is an important clinical task. However, there is a lack of a reliable, objective ground truth for evaluating automatic tracking methods. We have developed a simulation tool that can generate MR images with known tumor and edema. More... |
|
Evaluation of Registration AlgorithmsWe are interested in comparing existing registration packages to determine how registration in Slicer3 can be improved. This work focuses on examining various packages researchers are currently using for registration and comparing results on a set of examples representative of common registration tasks. More...
|
|
A Framework for Registration Evaluation and ExplorationIn response to the difficulties of image registration, we propose an environment where registration applications can be explored, tested, and compared. More... |