Difference between revisions of "BRAINSFit prostate registration"
| (2 intermediate revisions by the same user not shown) | |||
| Line 197: | Line 197: | ||
SimpleITK registration code on github: [https://github.com/PeterBehringer/SimpleITKtraining/blob/master/BRAINSFit_to_sitk_MMI.py here] | SimpleITK registration code on github: [https://github.com/PeterBehringer/SimpleITKtraining/blob/master/BRAINSFit_to_sitk_MMI.py here] | ||
| − | '''Latest update: Mai | + | '''Latest update: Mai 18, 2015''' |
==Things to do== | ==Things to do== | ||
| − | * Set | + | * Set ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS to 1 as described [http://comments.gmane.org/gmane.comp.lib.itk.user/44256 here] |
* Write message to ITK mailing list, cc Nicholas Tustin: ask for regularization of the BSpline displacement field | * Write message to ITK mailing list, cc Nicholas Tustin: ask for regularization of the BSpline displacement field | ||
* look at DICE comparison between Slicer3 and Slicer4 to quantify the problem | * look at DICE comparison between Slicer3 and Slicer4 to quantify the problem | ||
Latest revision as of 23:39, 18 May 2015
Home < BRAINSFit prostate registrationContents
Goal
We are developing registration module in Slicer version 4 (which is using ITKv4) for deformable registration of prostate MRI using BRAINSFit This Slicer4/ITKv4 registration tool should be functional, accurate and fast (i.e., comparable with the functionality we had in Slicer3/ITKv3, which has been evaluated previously [1]). We worked on implementing BRAINSFit registration in SimpleITK but due to several issues (see here) we are considering a C++ implementation that uses ITKv3 libraries and can be integrated in the latest Slicer versions.
Details on the registration approach
Registration is applied to align preprocedural and intraoperational MR T2 image volumes. We are using masks the prostate for both image data sets. Registration is done using MMI metric with rigid, affine and B-spline stages applied in sequence. In Slicer3/BRAINSFit we use gradient descent for rigid/affine, and LBFGS for B-spline.
Parameters we are using to call BRAINSFit in Slicer 3.6 and Slicer 4.4
| Parameter | Slicer 3.6 | Slicer 4.4 |
|---|---|---|
| --fixedVolume | dir | dir |
| --movingVolume | dir | dir |
| --outputVolume | dir | dir |
| --bsplineTransform | dir | dir |
| --movingBinaryVolume | dir | dir |
| --fixedBinaryVolume | dir | dir |
| --useCenterOfROIAlign | True | not an option |
| --initializeTransformMode | not an option | useCenterOfROIAlign |
| --samplingPercentage | not an option | 0.002 |
| --useRigid | True | True |
| --useAffine | True | True |
| --useROIBSpline | True | True |
| --useScaleVersor3D | True | True |
| --useScaleSkewVersor3D | True | True |
| --useBSpline | not an option | True |
| --splineGridSize | 3,3,3 | 3,3,3 |
| --numberOfIterations | 1500 | 1500 |
| --maskProcessing | ROI | ROI |
| --outputVolumePixelType | float | float |
| --backgroundFillValue | 0 | 0 |
| --maskInferiorCutOffFromCenter | 1000 | 1000 |
| --interpolationMode | Linear | Linear |
| --minimumStepSize | 0.005 | not an option |
| --minimumStepLength | not an option | 0.005 |
| --translationScale | 1000 | 1000 |
| --reproportionScale | 1 | 1 |
| --skewScale | 1 | 1 |
| --numberOfHistogramBins | 50 | 50 |
| --numberOfMatchPoints | 10 | 10 |
| --numberOfSamples | 100000 | not an option |
| --fixedVolumeTimeIndex | 0 | 0 |
| --movingVolumeTimeIndex | 0 | 0 |
| --medianFilterSize | 0,0,0 | 0,0,0 |
| --ROIAutoDilateSize | 0 | 0 |
| --relaxationFactor | 0.5 | 0.5 |
| --maximumStepSize | 0.2 | not an option |
| --maximumStepLength | not an option | 0.2 |
| --failureExitCode | -1 | -1 |
| --debugNumberOfThreads | -1 | not an option |
| --numberOfThreads | not an option | -1 |
| --debugLevel | 0 | 0 |
| --costFunctionConvergenceFactor | 1.00E+09 | 1.00E+09 |
| --projectedGradientTolerance | 1.00E-05 | 1.00E-05 |
| --maxBSplineDisplacement | 0 | 0 |
| --maximumNumberOfEvaluations | not an option | 900 |
| --maximumNumberOfCorrections | not an option | 25 |
| --metricSamplingStrategy | not an option | Random |
| --costMetric | not an option | MMI |
| --removeIntensityOutliers | not an option | 0
|
| --ROIAutoClosingSize | not an option | 9
|
| --useExplicitPDFDerivativesMode | AUTO | not an option |
| --useCachingOfBSplineWeightsMode | ON | not an option |
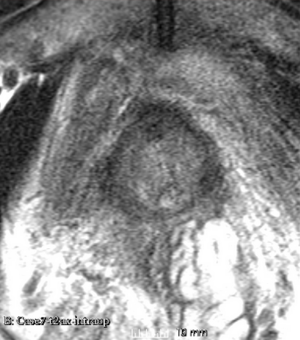
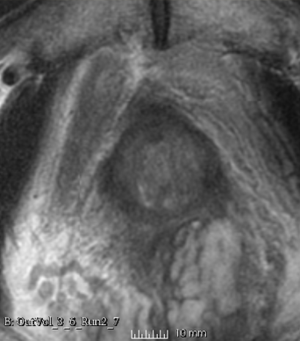
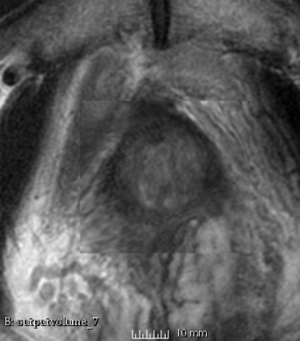
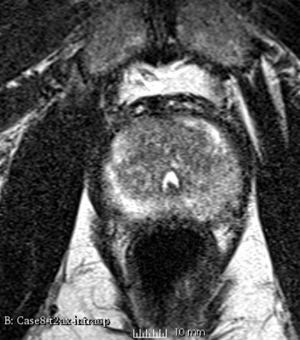
Results using the above parameters
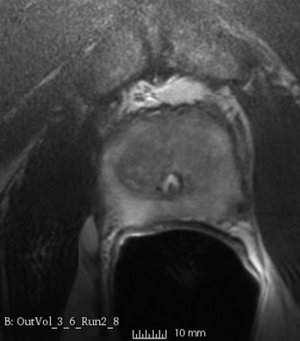
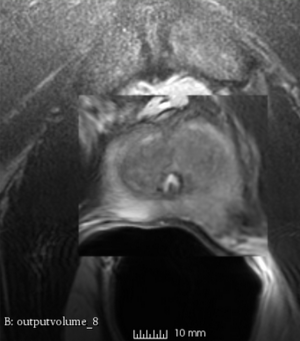
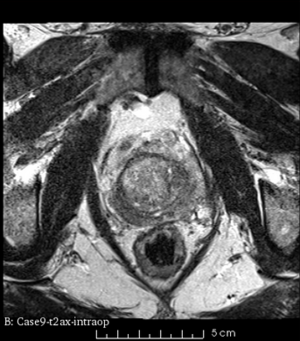
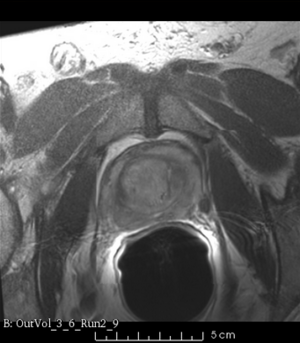
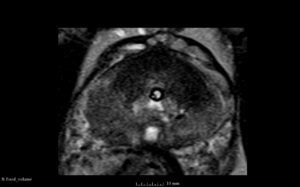
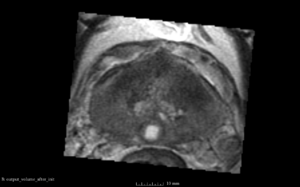
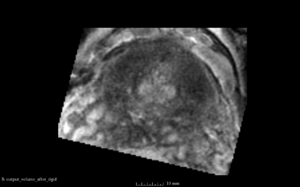
We used this sample data with Reader 1 segmentations to visually evaluate differences in the registration results. All three results of BRAINSFit with ITKv3 in Slicer 3.6 look reasonable. Using BRAINSFit with ITKv4 in Slicer 4.4 creates a reasonable result for Case 7, but look strongly distorted towards the edges of the bounding box in Case 8 and 9.
| Case 7 |
| Case 8 |
| Case 9 |
Current status
SimpleITK registration code on github: here
Latest update: Mai 18, 2015
Things to do
- Set ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS to 1 as described here
- Write message to ITK mailing list, cc Nicholas Tustin: ask for regularization of the BSpline displacement field
- look at DICE comparison between Slicer3 and Slicer4 to quantify the problem
- displace the masks and run with Slicer3 and Slicer4 to quantify the impact of the mask in the registration result
Things that are working
- exhaustive search based initialization procedure implemented and produces good results (cross-correlation metric) in SimpleITK
Things that are implemented, but are not working
- SimpleITK code for rigid registration
- the voxel sampling exception can be avoided by cropping the images sharply around the masks.
- The rigid registration result is too much rotated and translated. Also the number of iteration steps varies with every computation. Reason might be that seed is set randomized and not set as done in BRAINSFit here. There is no option at the SimpleITK::ImageRegistrationMethod to set a metric seed.
Things that are not implemented, but need to be implemented
- SimpleITK code for affine registration
- SimpleITK code for BSpline registration (started here)
Unresolved issues out of our direct control
Standing SimpleITK issues
- no API for consistent initialization of the metric seed
- The ITKv4 ImageRegistrationMethod random sampling utilizes a fixed seed (here)
Standing ITKv4 issues
- capability to sample voxels only within the mask (see topic under review on Gerrit: http://review.source.kitware.com/#/c/19684/)
Standing Slicer4 issues
References
[1] Fedorov A, Tuncali K, Fennessy FM, Tokuda J, Hata N, et al. (2012) Image registration for targeted MRI-guided transperineal prostate biopsy. J Magn Reson Imaging 36: 987–992. Available: http://dx.doi.org/10.1002/jmri.23688.