Difference between revisions of "DBP2:UNCFinal:2010"
From NAMIC Wiki
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (29 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
[[DBP2:Main|back to DBP2 Main]] | [[DBP2:Main|back to DBP2 Main]] | ||
| + | __NOTOC__ | ||
| + | =Overview= | ||
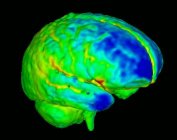
| + | [[Image:BrainDevelopment.jpg|thumb|177px|Brain development]] | ||
| − | + | The analysis of neuroimaging data from pediatric populations presents several challenges. Normal variations in brain shape exist from infancy to adulthood as well as normal developmental changes related to tissue maturation. Measuring cortical thickness is one important way to analyze such developmental tissue changes. As a DBP, our group is interested in the longitudinal study of early brain development as reflected in variations in cortical thickness in autistic children and controls. | |
| + | *We first developed a novel framework to perform individual regional cortical thickness analysis [[{{fullurl:{{FULLPAGENAME}}}}#ref_2 2], [{{fullurl:{{FULLPAGENAME}}}}#ref_4 4], [{{fullurl:{{FULLPAGENAME}}}}#ref_5 5]]. This framework relies first on atlas-based automatic tissue segmentation via an expectation maximization scheme to compute probabilistic and hard tissue segmentations. Second, the skull is stripped by using a post-processed hard tissue segmentation label map as a mask. Third, a B-spline registration, initialized by centers of mass, rigid then affine registration, is computed to register a T1-weighted atlas to the corrected T1w skull-stripped image. A multi-threaded coarse-to-fine registration scheme using mattes mutual information metric is considered. The full transformation is then applied to a lobar parcellation map defined in the atlas space, dividing the brain into 20+ lobes. By using both the tissue segmentation label map and the registered parcellation map, sparse and asymmetric cortical thickness measurements are finally computed for each lobe. | ||
| + | **This framework has been incorporated in our ARCTIC tool, an open source C++ based high-level module disseminated as part of the 3D Slicer toolkit. ARCTIC’s setup allows for efficient batch processing and grid computing via BatchMake (Kitware, Inc.). | ||
| + | **We performed a longitudinal MRI study to investigate early growth trajectories in brain volume and cortical thickness. Cerebral gray and white matter volumes and cortical thickness in children with autism spectrum disorder and controls were examined. Subjects were seen at approximately 2 years of age (autism = 59, controls = 38) and then were rescanned approximately 24 months later at age 4-5 years (autism = 38, controls = 21). Results and conclusions should be published soon (paper submitted to AGP, Archives of General Psychiatry). | ||
| − | + | *We also developed a novel framework that permits group-wise automatic mesh-based analysis of cortical thickness [[{{fullurl:{{FULLPAGENAME}}}}#ref_1 1], [{{fullurl:{{FULLPAGENAME}}}}#ref_3 3], [{{fullurl:{{FULLPAGENAME}}}}#ref_6 6], [{{fullurl:{{FULLPAGENAME}}}}#ref_7 7]]. Our analysis framework consists of a pipeline of C++ based automated 3D Slicer compatible modules. The approach is divided into four parts. First an individual pre-processing pipeline is applied on each subject to create genus-zero inflated white matter cortical surfaces with cortical thickness measurements. The second part performs an entropy-based group-wise shape correspondence on these meshes using a particle system, which establishes a trade-off between an even sampling of the cortical surfaces and the similarity of corresponding points across the population using sulcal depth information and spatial proximity. A novel automatic initial particle sampling is performed using a matched 98-lobe parcellation map prior to a particle-splitting phase. Third, corresponding re-sampled surfaces are computed with interpolated cortical thickness measurements, which are finally analyzed via a statistical vertex-wise analysis module. | |
| + | **This framework has been tested on a small pediatric dataset and incorporated in an open source C++ based high-level module called GAMBIT. GAMBIT’s setup allows efficient batch processing, grid computing and quality control. | ||
=Software= | =Software= | ||
| − | *ARCTIC | + | [[Image:ArcticLogo.png|thumb|150px|ARCTIC Logo|right]] |
| + | *[http://www.nitrc.org/projects/gambit ARCTIC] (Automatic Regional Cortical ThICkness) | ||
**Description: | **Description: | ||
***ARCTIC is an end-to-end application allowing individual lobar analysis of cortical thickness | ***ARCTIC is an end-to-end application allowing individual lobar analysis of cortical thickness | ||
| − | ***Pipeline: | + | ***Pipeline: tissue segmentation, regional atlas deformable registration, cortical thickness measurements, volume information stored in spreadsheets |
***Visualization: white matter and gray matter mesh creation | ***Visualization: white matter and gray matter mesh creation | ||
***Quality control: optimal QC via 3D Slicer MRML scenes | ***Quality control: optimal QC via 3D Slicer MRML scenes | ||
**Download: | **Download: | ||
| + | ***Source code, executables and tutorial are available on [http://www.nitrc.org/projects/gambit NITRC] | ||
***Latest stable version is directly available as an extension in [http://www.slicer.org/pages/Special:SlicerDownloads Slicer 3.6.1 release] and soon in Slicer 3.6.2 | ***Latest stable version is directly available as an extension in [http://www.slicer.org/pages/Special:SlicerDownloads Slicer 3.6.1 release] and soon in Slicer 3.6.2 | ||
| − | |||
**Documentation: | **Documentation: | ||
***[http://www.nitrc.org/plugins/mwiki/index.php/arctic:MainPage NITRC wiki page] | ***[http://www.nitrc.org/plugins/mwiki/index.php/arctic:MainPage NITRC wiki page] | ||
| − | ***[ | + | ***[https://www.slicer.org/wiki/Modules:ARCTIC-Documentation-3.6 Online documentation within Slicer 3.6] |
**Tutorials: | **Tutorials: | ||
***ARCTIC tutorials: [[Media:ARCTIC-Slicer3-Tutorial.ppt| [ppt]]][[Media:ARCTIC-Slicer3-Tutorial.pdf| [pdf]]] | ***ARCTIC tutorials: [[Media:ARCTIC-Slicer3-Tutorial.ppt| [ppt]]][[Media:ARCTIC-Slicer3-Tutorial.pdf| [pdf]]] | ||
****1st Prize: NAMIC tutorial contest AHM 2009 | ****1st Prize: NAMIC tutorial contest AHM 2009 | ||
****2nd Prize: NAMIC tutorial contest summer project week 2009 | ****2nd Prize: NAMIC tutorial contest summer project week 2009 | ||
| − | |||
| − | |||
| + | <center> | ||
| + | {|class=wikitable | ||
| + | |[[Image:T1Image.jpg|150px|T1-weighted skull-stripped image]] | ||
| + | |[[Image:Parcellation.jpg|150px|Parcellation image]] | ||
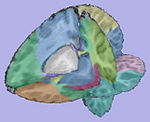
| + | |[[Image:WMThickness.jpg|150px|Cortical thickness on WM surface]] | ||
| + | |[[Image:ThicknessInformation.jpg|150px|Cortical thickness information]] | ||
| + | |- | ||
| + | |T1-weighted skull-stripped image | ||
| + | |Parcellation image | ||
| + | |Cortical thickness on WM surface | ||
| + | |Cortical thickness information | ||
| + | |} | ||
| + | </center> | ||
| − | *GAMBIT | + | [[Image:GAMBITLogo.png|thumb|150px|GAMBIT Logo|right]] |
| + | *[http://www.nitrc.org/projects/gambit GAMBIT] (Group-wise Automatic Mesh Based analysis of cortIcal Thickness) | ||
**Description: | **Description: | ||
***GAMBIT is an end-to-end application allowing allowing group-wise automatic mesh-based analysis of cortical thickness as well as other surface area measurements | ***GAMBIT is an end-to-end application allowing allowing group-wise automatic mesh-based analysis of cortical thickness as well as other surface area measurements | ||
| Line 33: | Line 53: | ||
***Quality control: optimal QC via 3D Slicer MRML scenes | ***Quality control: optimal QC via 3D Slicer MRML scenes | ||
**Download: | **Download: | ||
| + | ***Source code, executables and tutorial are available on [http://www.nitrc.org/projects/gambit NITRC] | ||
***Latest stable version is available soon as an extension in Slicer 3.6.2 | ***Latest stable version is available soon as an extension in Slicer 3.6.2 | ||
| − | |||
**Documentation: | **Documentation: | ||
***[http://www.nitrc.org/plugins/mwiki/index.php/gambit:MainPage NITRC wiki page] | ***[http://www.nitrc.org/plugins/mwiki/index.php/gambit:MainPage NITRC wiki page] | ||
**Tutorials: | **Tutorials: | ||
***GAMBIT tutorial presentations: [http://wiki.na-mic.org/Wiki/index.php/File:GAMBIT_TutorialContestSummer2010.ppt [ppt]] [http://wiki.na-mic.org/Wiki/index.php/File:GAMBIT_TutorialContestSummer2010.pdf [pdf]] | ***GAMBIT tutorial presentations: [http://wiki.na-mic.org/Wiki/index.php/File:GAMBIT_TutorialContestSummer2010.ppt [ppt]] [http://wiki.na-mic.org/Wiki/index.php/File:GAMBIT_TutorialContestSummer2010.pdf [pdf]] | ||
| − | + | ||
| + | <center> | ||
| + | {|class=wikitable | ||
| + | |[[Image:GAMBIT QC CorticalThickness.png|500px|Cortical thickness overlayed on inflated cortical surfaces]] | ||
| + | |valign="center"|[[Image:GAMBIT_QC_SulcalDepth.png|500px|Sucal depth overlayed on inflated cortical surfaces]] | ||
| + | |- | ||
| + | |Cortical thickness overlayed on inflated cortical surfaces | ||
| + | |Sucal depth overlayed on inflated cortical surfaces | ||
| + | |} | ||
| + | </center> | ||
=Listing and short description of the sample data= | =Listing and short description of the sample data= | ||
| − | *Pediatric Brain MRI data | + | [[Image:MeshBasedCortThick_T1Image.jpg|thumb|125px|T1-weighted image]] |
| + | *Pediatric Brain MRI data available on MIDAS | ||
**[http://insight-journal.org/midas/community/view/24 Data of 2 autistic children and 2 normal controls] (male, female) scanned at 2 years with follow up at 4 years from a 1.5T Siemens scanner. Files include structural data, tissue segmentation label map and subcortical structures segmentation. | **[http://insight-journal.org/midas/community/view/24 Data of 2 autistic children and 2 normal controls] (male, female) scanned at 2 years with follow up at 4 years from a 1.5T Siemens scanner. Files include structural data, tissue segmentation label map and subcortical structures segmentation. | ||
| − | + | [[Image:MeshBasedCortThick_BrainROIAtlas_AllROIMesh.jpg|thumb|125px|T1-weigthed atlas with subcortical structures]] | |
| − | + | *Brain Atlases available on MIDAS | |
| − | + | ** Average T1-weighted images (with/without skull) are provided with tissue segmentation probability maps (white matter, gray matter, csf, rest), subcortical structures probability maps (amygdala, caudate, hippocampus, pallidus, putamen) and lobar parcellation maps | |
| − | |||
| − | *Brain Atlases | ||
**[http://www.insight-journal.org/midas/item/view/2277 Pediatric atlas] | **[http://www.insight-journal.org/midas/item/view/2277 Pediatric atlas] | ||
**[http://www.insight-journal.org/midas/item/view/2328 Adult atlas] | **[http://www.insight-journal.org/midas/item/view/2328 Adult atlas] | ||
**[http://www.insight-journal.org/midas/item/view/2330 Elderly atlas] | **[http://www.insight-journal.org/midas/item/view/2330 Elderly atlas] | ||
**[http://www.insight-journal.org/midas/item/view/2283 Primate atlas] | **[http://www.insight-journal.org/midas/item/view/2283 Primate atlas] | ||
| + | *Tutorial datasets: | ||
| + | **[http://www.nitrc.org/frs/download.php/545/ARCTIC_Tutorial_example_1.0.zip Download ARCTIC tutorial dataset] | ||
| + | **[http://www.nitrc.org/frs/download.php/2185/GAMBIT_Tutorial_example_1.0.zip Download GAMBIT tutorial dataset] | ||
=Related pages= | =Related pages= | ||
*[http://www.slicer.org/pages/Special:SlicerDownloads Slicer 3.6 download] | *[http://www.slicer.org/pages/Special:SlicerDownloads Slicer 3.6 download] | ||
| − | *[ | + | *[https://www.slicer.org/wiki/Documentation-3.6 Slicer 3.6 documentation] |
| + | *[http://www.niral.unc.edu/ Neuro Image Research and Analysis Laboratory, UNC Chapel Hill] | ||
| + | *[http://www.cidd.unc.edu/ UNC Carolina Institute for Developmental Disabilities] | ||
=References= | =References= | ||
| − | + | #<cite id="ref_1"></cite>C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Group-wise Automatic Mesh-Based Analysis of Cortical Thickness, Proc. SPIE 7962, 796227 (2011) | |
| − | + | #<cite id="ref_2"></cite>Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, Vachet, C., & Piven, J., Early brain overgrowth in autism results from an increase in cortical surface area before age 2, Arch Gen Psychiatry. 2011;68(5):1-10 | |
| + | #<cite id="ref_3"></cite>Oguz I., Niethammer M., Cates J., Whitaker R., Fletcher T., Vachet C., Styner M. [http://www.na-mic.org/publications/item/view/1671 Cortical Correspondence with Probabilistic Fiber Connectivity.] Inf Process Med Imaging. 2009;21:651-63. PMID: 19694301. PMCID: PMC2751643. | ||
| + | #<cite id="ref_4"></cite>H.C. Hazlett, C. Vachet, C. Mathieu, M. Styner, J. Piven, Use of the Slicer3 Toolkit to Produce Regional Cortical Thickness Measurement of Pediatric MRI Data, presented at the 8th Annual International Meeting for Autism Research (IMFAR) Chicago, IL 2009. | ||
| + | #<cite id="ref_5"></cite>C. Mathieu, C. Vachet, H.C. Hazlett, G. Geric, J. Piven, and M. Styner, ARCTIC – Automatic Regional Cortical ThICkness Tool, UNC Radiology Research Day 2009 abstract. | ||
| + | #<cite id="ref_6"></cite>Oguz, I., Cates, J., Fletcher, T., Whitaker, R., Cool, D., Aylward, S., Styner, M., Cortical correspondence using entropy-based particle systems and local features, IEEE Symposium on Biomedical Imaging ISBI 2008. 1637-1640. | ||
| + | #<cite id="ref_7"></cite>J. Cates J., Fletcher P.T., Styner M., Hazlett H.C., Whitaker R. [http://www.na-mic.org/publications/item/view/1473 Particle-Based Shape Analysis of Multi-object Complexes.] Int Conf Med Image Comput Comput Assist Interv. 2008;11(Pt 1):477-485. PMID: 18979781. PMCID: PMC2753605. | ||
Latest revision as of 18:07, 10 July 2017
Home < DBP2:UNCFinal:2010back to DBP2 Main
Overview
The analysis of neuroimaging data from pediatric populations presents several challenges. Normal variations in brain shape exist from infancy to adulthood as well as normal developmental changes related to tissue maturation. Measuring cortical thickness is one important way to analyze such developmental tissue changes. As a DBP, our group is interested in the longitudinal study of early brain development as reflected in variations in cortical thickness in autistic children and controls.
- We first developed a novel framework to perform individual regional cortical thickness analysis [2, 4, 5]. This framework relies first on atlas-based automatic tissue segmentation via an expectation maximization scheme to compute probabilistic and hard tissue segmentations. Second, the skull is stripped by using a post-processed hard tissue segmentation label map as a mask. Third, a B-spline registration, initialized by centers of mass, rigid then affine registration, is computed to register a T1-weighted atlas to the corrected T1w skull-stripped image. A multi-threaded coarse-to-fine registration scheme using mattes mutual information metric is considered. The full transformation is then applied to a lobar parcellation map defined in the atlas space, dividing the brain into 20+ lobes. By using both the tissue segmentation label map and the registered parcellation map, sparse and asymmetric cortical thickness measurements are finally computed for each lobe.
- This framework has been incorporated in our ARCTIC tool, an open source C++ based high-level module disseminated as part of the 3D Slicer toolkit. ARCTIC’s setup allows for efficient batch processing and grid computing via BatchMake (Kitware, Inc.).
- We performed a longitudinal MRI study to investigate early growth trajectories in brain volume and cortical thickness. Cerebral gray and white matter volumes and cortical thickness in children with autism spectrum disorder and controls were examined. Subjects were seen at approximately 2 years of age (autism = 59, controls = 38) and then were rescanned approximately 24 months later at age 4-5 years (autism = 38, controls = 21). Results and conclusions should be published soon (paper submitted to AGP, Archives of General Psychiatry).
- We also developed a novel framework that permits group-wise automatic mesh-based analysis of cortical thickness [1, 3, 6, 7]. Our analysis framework consists of a pipeline of C++ based automated 3D Slicer compatible modules. The approach is divided into four parts. First an individual pre-processing pipeline is applied on each subject to create genus-zero inflated white matter cortical surfaces with cortical thickness measurements. The second part performs an entropy-based group-wise shape correspondence on these meshes using a particle system, which establishes a trade-off between an even sampling of the cortical surfaces and the similarity of corresponding points across the population using sulcal depth information and spatial proximity. A novel automatic initial particle sampling is performed using a matched 98-lobe parcellation map prior to a particle-splitting phase. Third, corresponding re-sampled surfaces are computed with interpolated cortical thickness measurements, which are finally analyzed via a statistical vertex-wise analysis module.
- This framework has been tested on a small pediatric dataset and incorporated in an open source C++ based high-level module called GAMBIT. GAMBIT’s setup allows efficient batch processing, grid computing and quality control.
Software
- ARCTIC (Automatic Regional Cortical ThICkness)
- Description:
- ARCTIC is an end-to-end application allowing individual lobar analysis of cortical thickness
- Pipeline: tissue segmentation, regional atlas deformable registration, cortical thickness measurements, volume information stored in spreadsheets
- Visualization: white matter and gray matter mesh creation
- Quality control: optimal QC via 3D Slicer MRML scenes
- Download:
- Source code, executables and tutorial are available on NITRC
- Latest stable version is directly available as an extension in Slicer 3.6.1 release and soon in Slicer 3.6.2
- Documentation:
- Tutorials:
- Description:
|
|
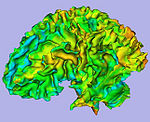
|
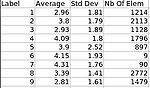
|
| T1-weighted skull-stripped image | Parcellation image | Cortical thickness on WM surface | Cortical thickness information |
- GAMBIT (Group-wise Automatic Mesh Based analysis of cortIcal Thickness)
- Description:
- GAMBIT is an end-to-end application allowing allowing group-wise automatic mesh-based analysis of cortical thickness as well as other surface area measurements
- Pipeline: individual preprocessing pipeline, group-wise particle-based correspondence on inflated genus-zero white matter surfaces, group-wise statistical analysis
- Visualization: inflated and folded white matter surfaces in correspondence, with cortical thickness and sulcal depth as overlays
- Quality control: optimal QC via 3D Slicer MRML scenes
- Download:
- Source code, executables and tutorial are available on NITRC
- Latest stable version is available soon as an extension in Slicer 3.6.2
- Documentation:
- Tutorials:
- Description:

|
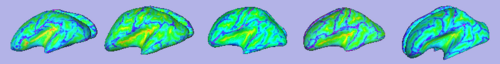
|
| Cortical thickness overlayed on inflated cortical surfaces | Sucal depth overlayed on inflated cortical surfaces |
Listing and short description of the sample data
- Pediatric Brain MRI data available on MIDAS
- Data of 2 autistic children and 2 normal controls (male, female) scanned at 2 years with follow up at 4 years from a 1.5T Siemens scanner. Files include structural data, tissue segmentation label map and subcortical structures segmentation.
- Brain Atlases available on MIDAS
- Average T1-weighted images (with/without skull) are provided with tissue segmentation probability maps (white matter, gray matter, csf, rest), subcortical structures probability maps (amygdala, caudate, hippocampus, pallidus, putamen) and lobar parcellation maps
- Pediatric atlas
- Adult atlas
- Elderly atlas
- Primate atlas
- Tutorial datasets:
Related pages
- Slicer 3.6 download
- Slicer 3.6 documentation
- Neuro Image Research and Analysis Laboratory, UNC Chapel Hill
- UNC Carolina Institute for Developmental Disabilities
References
- C. Vachet, H.C. Hazlett, M. Niethammer, I. Oguz, J.Cates, R. Whitaker, J. Piven, M. Styner, Group-wise Automatic Mesh-Based Analysis of Cortical Thickness, Proc. SPIE 7962, 796227 (2011)
- Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, Vachet, C., & Piven, J., Early brain overgrowth in autism results from an increase in cortical surface area before age 2, Arch Gen Psychiatry. 2011;68(5):1-10
- Oguz I., Niethammer M., Cates J., Whitaker R., Fletcher T., Vachet C., Styner M. Cortical Correspondence with Probabilistic Fiber Connectivity. Inf Process Med Imaging. 2009;21:651-63. PMID: 19694301. PMCID: PMC2751643.
- H.C. Hazlett, C. Vachet, C. Mathieu, M. Styner, J. Piven, Use of the Slicer3 Toolkit to Produce Regional Cortical Thickness Measurement of Pediatric MRI Data, presented at the 8th Annual International Meeting for Autism Research (IMFAR) Chicago, IL 2009.
- C. Mathieu, C. Vachet, H.C. Hazlett, G. Geric, J. Piven, and M. Styner, ARCTIC – Automatic Regional Cortical ThICkness Tool, UNC Radiology Research Day 2009 abstract.
- Oguz, I., Cates, J., Fletcher, T., Whitaker, R., Cool, D., Aylward, S., Styner, M., Cortical correspondence using entropy-based particle systems and local features, IEEE Symposium on Biomedical Imaging ISBI 2008. 1637-1640.
- J. Cates J., Fletcher P.T., Styner M., Hazlett H.C., Whitaker R. Particle-Based Shape Analysis of Multi-object Complexes. Int Conf Med Image Comput Comput Assist Interv. 2008;11(Pt 1):477-485. PMID: 18979781. PMCID: PMC2753605.