DBP:Harvard:Software:Testing:EMABC Validation
Contents
Validating Slicer Module 'EMAtlasBrainClassifier'
Katharina Quintus, PNL
Introduction
1.1 Goals:
Studies performed by the Psychiatry Neuroimaging Laboratory (PNL) rely on automatic segmentation of gray matter, white matter and cerebrospinal fluid (CSF) from brain MR images. We are interested in a segementation pipeline that (i) requires minimal human interaction, and is (ii) easy to maintain and control from a technical point of view. In the following the results of such a new method are compared to the results from our current segementation pipeline.
1.2. Current segmentation pipeline:
So far for every scanned volume involved in studies has been segmented into white matter, gray matter and CSF using the following four step segmentation pipeline:
- Coregistration of T2 and structural volumes using the Slicer “AG” Module, which transforms and re-slices the T2 volume so that it lines up with the SPGR.
- Intensity Normalization: Scaling the intensity of every scan to match its average intensity to a template in order to adjust for different intensity profiles resulting from different scanners used. This is done using the Normalization module in Slicer.
- Atlas Registration: Warping a template atlas to the case being segmented which yields four probability atlases (one for every tissue class, one for background). Currently this is performed by a Python script written by Alexandre Guimond.
- EM Segmentation: The expectation maximization procedure separates MR data into background, face, CSF, gray matter, and white matter. Up to now this is accomplished by a Tcl script written by Kilian Pohl.
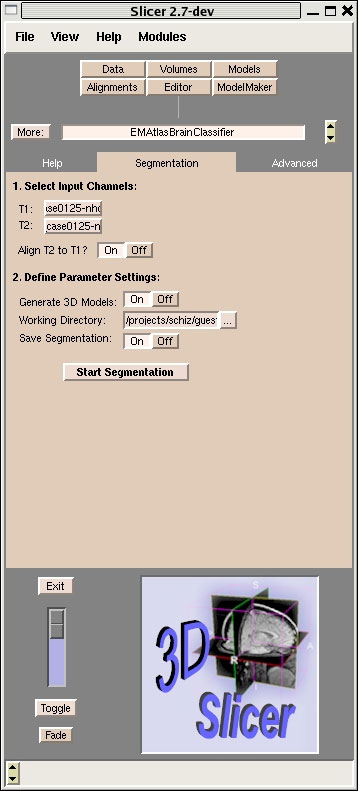
1.3. New pipeline: The EMAtlasBrainClassifier module in Slicer:
The EMAtlasBrainClassifier module basically implements the same expectation maximization algorithm as the Tcl script used in the current PNL processing pipeline. However, this module performs all four processing steps with a single button press. The user has to define the structural volume and the corresponding T2 volume and decides if step one (coregistration) is needed or not. Furthermore the user can turn on/off the output of various intermediate results of the segmentation algorithm under ”Advanced Tab”. Figure 1 shows a screenshot of the user interface of the EMAtlasBrainClassifier Module in Slicer.