Difference between revisions of "Projects/Structural/2007 Project Week EMSegmentation Validation"
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:ProjectWeek-2007.png|thumb|320px|Return to [[2007_Programming/Project_Week_MIT|Project Week Main Page]] ]] | |[[Image:ProjectWeek-2007.png|thumb|320px|Return to [[2007_Programming/Project_Week_MIT|Project Week Main Page]] ]] | ||
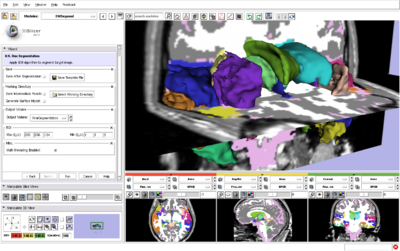
| − | |[[Image:EMSegment31Structures.png|thumb| | + | |[[Image:EMSegment31Structures.png|thumb|400px|Brain segmentation using EMSegment and ModelMaker in Slicer3.]] |
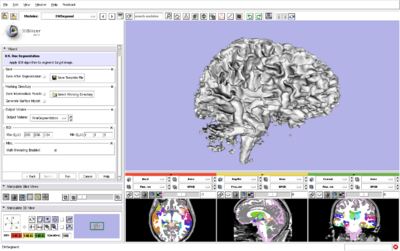
| + | |[[Image:EMSegment31Structures2.png|thumb|400px|Brain segmentation using EMSegment and ModelMaker in Slicer3.]] | ||
|} | |} | ||
| Line 30: | Line 31: | ||
Preparations: develop EMSegment command line executable (DONE), add intensity normalization to module (Done) | Preparations: develop EMSegment command line executable (DONE), add intensity normalization to module (Done) | ||
| + | |||
| + | Discussions: registration in EMSegmenter (design, coding UNDERWAY), application to white matter lesions (Jeremy Bockholt, Mark Scully at MIND, Kilian Pohl) | ||
| + | |||
| + | Updates: allow user to specify atlas files from the command line (DONE) | ||
| + | |||
| + | To Do: begin validation study | ||
</div> | </div> | ||
Latest revision as of 13:06, 29 June 2007
Home < Projects < Structural < 2007 Project Week EMSegmentation Validation Return to Project Week Main Page |
Key Investigators
- Kitware: Brad Davis
- BWH: Sylvain Bouix
Objective
Our goal is to begin a validation study that will establish the accuracy and dependability of the Slicer3 EMSegment module. This objective is part of a larger, ongoing development effort for this module.
Approach, Plan
The validation study is based on a statistical comparison with results from earlier implementations of the EMSegment algorithm. The study will be carried out by members of the Psychiatry Neuroimiging Laboratory (PNL) at Brigham and Women's Hospital, on their data, using the newly developed command line version of the EMSegment algorithm. Kitware will provide software support and assist if problems arise with the results. Details are provided in the reference below. Our plan for the project week is to train the PNL staff on use of the new EMSegment tools, coordinate the validation study, and work out any bugs/interface problems as early as possible.
Progress
Preparations: develop EMSegment command line executable (DONE), add intensity normalization to module (Done)
Discussions: registration in EMSegmenter (design, coding UNDERWAY), application to white matter lesions (Jeremy Bockholt, Mark Scully at MIND, Kilian Pohl)
Updates: allow user to specify atlas files from the command line (DONE)
To Do: begin validation study