Difference between revisions of "Projects:PathologyAnalysis"
| (128 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
| − | = | + | = Ongoing Work = |
| − | == | + | == 4D Active Cut: An Interactive Tool for Pathological Anatomy Modeling == |
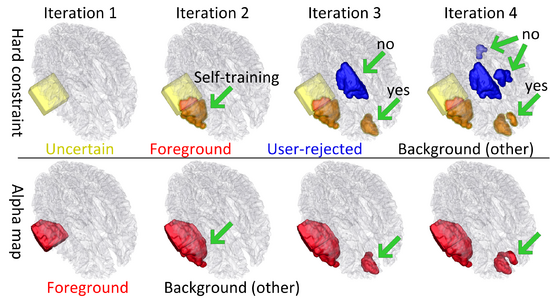
| − | + | We propose a novel semi-supervised method, called 4D active cut for lesion recognition and deformation estimation. Existing interactive segmentation methods passively wait for user to refine the segmentations which is a difficult task in 3D images that change over time. 4D active cut instead actively selects candidate regions for querying the user, and therefore obtains the most informative user feedback. A user simply answers `yes' or `no' to a candidate object without having to refine the segmentation slice by slice. Compared to single-object detection of GrabCut, our method also detects multiple lesions with spatial coherence using Markov random fields constraints. [1] | |
| − | + | [[File:Flowchart_4D_active_cut.png|450px|center|thumb|Flowchart of 4D active cut framework.]] | |
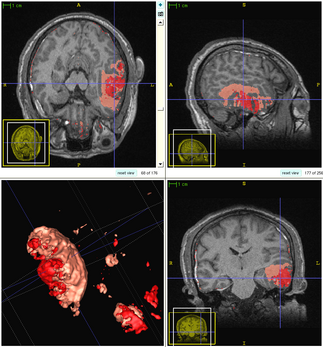
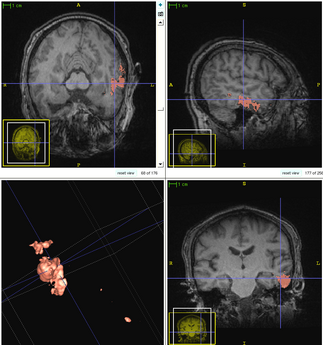
| − | + | The following figure shows the dynamic learning process of the proposed framework applied to one subject. | |
| − | |||
| + | [[File:Overview_4D_active_cut.png|550px|center|thumb|llustration of the iterative process of 4D active cut on one subject. Iteration 1: User-initialized bounding box. Iteration 2: Self training. Iteration 3 and 4: Active learning. The user stopped the process at 5th iteration. Arrows point to the changes between iterations. The white matter surface is visualized in all figures as reference.]] | ||
| + | |||
| + | == Modeling 4D Changes in Pathological Anatomy == | ||
| + | |||
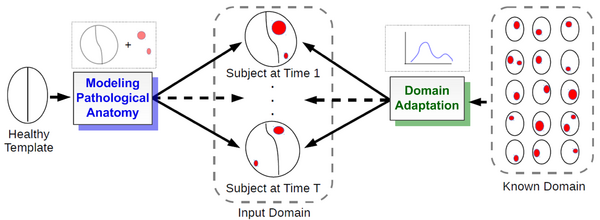
| + | We propose a novel framework that models 4D changes in pathological anatomy across time, and provides explicit mapping from a healthy template to subjects with pathology. Moreover, our framework uses transfer learning to leverage rich information from a known source domain, where we have a collection of completely segmented images, to yield effective appearance models for the input target domain. The automatic 4D segmentation method uses a novel domain adaptation technique for generative kernel density models to transfer information between different domains, resulting in a fully automatic method that requires no user interaction. [2] | ||
| + | |||
| + | [[File:PAM_concept_figure.png|600px|center|thumb|Conceptual overview of the proposed framework.]] | ||
| + | |||
| + | == Analyzing imaging biomarkers for traumatic brain injury using 4D modeling of longitudinal MRI == | ||
| + | <!-- During the NAMIC project week in Boston, we had a discussion with our collaborators in UCLA about the computation and visualization of cortical atrophy and tissue deformation. We want to visualize the cortical atrophy and tissue deformation at the same time. The following are some initial result. | ||
| + | |||
| + | [[File:cortical_atrophy_image.png|400px|center|thumb|Preliminary result of cortical atrophy computation and visualization]] | ||
| + | |||
| + | --> | ||
| + | |||
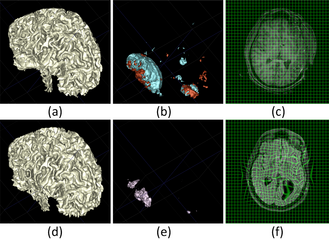
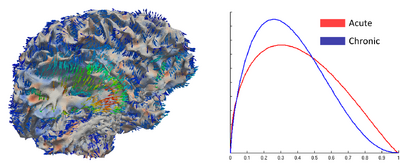
| + | We present a new method for computing surface-based and voxel-based imaging biomarkers using 4D modeling of longitudinal MRI. We analyze the potential for clinical use of these biomarkers by correlating them with TBI-specific patient scores. Our preliminary results indicate that the proposed voxel-based biomarkers are correlated with clinical outcomes [3]. | ||
| + | |||
| + | <center> | ||
| + | <gallery widths=400px heights=240px perrow=2 > | ||
| + | File:Result4DModeling.png|Spatiotemporal model for a subject with severe TBI. Top: 3D display of WM (a), lesions (b) and (c) regular grid for the acute time point. Bottom: Same displays for chronic time point (d) and (e), and deformation mapping from actue to chronic shown in (f). | ||
| + | File:cortical_atrophy_image.png| Left: Temporal change of cortical thickness (color) shown with tissue deformation (arrows). Right: Cortical thickness distribution of whole brain from acute to chronic. | ||
| + | |||
| + | </gallery> | ||
| + | </center> | ||
| + | |||
| + | == Segmentation of Serial MRI of TBI patients using Personalized Atlas Construction and Topological Change Estimation == | ||
| + | |||
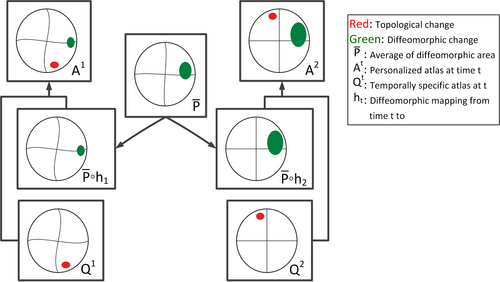
| + | Longitudinal images with TBI present topological changes over time due to the effect of the impact force on tissue, skull, and blood vessels and the recovery process. We address this issue by defining a novel atlas construction scheme that explicitly models the effect of topological changes [4]. Our method automatically estimates the probability of topological changes jointly with the personalized atlas. The following figure shows our atlas construction scheme. | ||
| + | |||
| + | |||
| + | [[File:Atlas_contruction_illu_TBI.png|500px|center|thumb|Construction of a personalized spatiotemporal atlas | ||
| + | using diffeomorphic and non-diffeomorphic components.]] | ||
| + | Our method combines 4D information through the creation of personalized | ||
| + | atlas that explicitly handles diffeomorphic and nondiffeomorphic | ||
| + | temporal changes. The method is robust to | ||
| + | topological changes caused by the injury and the recovery | ||
| + | process in TBI. | ||
| + | |||
| + | == Patient-Specific Segmentation Framework for Longitudinal MR Images of Traumatic Brain Injury == | ||
| + | |||
| + | We propose a multi-modal image segmentation framework for longitudinal TBI images [5]. The framework is initialized through manual input of primary lesion sites at each time point, which are then refined by a joint approach composed of Bayesian segmentation and construction of a personalized atlas. The personalized atlas construction estimates the average of the posteriors of the Bayesian segmentation at each time point and warps the average back to each time point to provide the updated priors for Bayesian segmentation. The difference between our approach and segmenting longitudinal images independently is that we use the information from all time points to improve the segmentations. Given a manual initialization, our framework automatically segments healthy structures (white matter, grey matter, cerebrospinal fluid) as well as different lesions such as hemorrhagic lesions and edema. Our framework can handle different sets of modalities at each time point, which provides flexibility in analyzing clinical scans. | ||
| + | |||
| + | === Segmentation framework === | ||
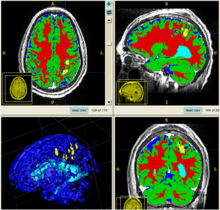
| + | The segmentation framework is initialized through manual input of primary lesion sites at each time point, which are then refined by a joint approach composed of Bayesian segmentation and construction of a personalized atlas. The following figure is the segmentation framework for multi-modal MR images. | ||
| + | |||
| + | [[File:Seg_algorithm_framework.png|550px|center|thumb|Segmentation framework]] | ||
| + | |||
| + | === Construction of personalized atlas === | ||
| + | Based on the segmentation results (the posteriors), we create personalized atlases using the unbiased diffeomorphic atlas construction method. In atlas construction, we estimate an average that requires the minimum amount of deformation to transform into the posteriors at every time point. The following flowchart shows the estimation of personalized atlases. | ||
| + | |||
| + | [[File:Joint_analysis_flowchart.png|450px|center|thumb|Flowchart of the estimation of personalized atlases]] | ||
| + | |||
| + | === Results === | ||
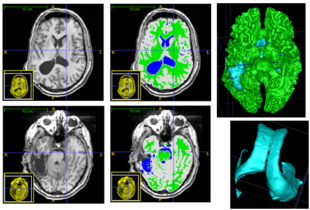
| + | Using the deformation field got from the estimation of personalized atlases, we can visualize the tissue deformation. In the following two figures, the deformation field is visualized as vector magnitude (left) and Jacobian (right). | ||
| + | |||
| + | <center> | ||
{| border="0" style="background:transparent;" | {| border="0" style="background:transparent;" | ||
| − | |[[ | + | |[[File:Vector mag 2D.png|300px|thumb|Visualization of vector magnitude]] |
| + | |[[File:Jacobian det 2D.png|300px|thumb|Visualization of Jacobian]] | ||
|- | |- | ||
|} | |} | ||
| + | </center> | ||
| + | The following two figures show the snapshot of the segmentation result. | ||
| − | + | <center> | |
| − | |||
| − | |||
| − | |||
{| border="0" style="background:transparent;" | {| border="0" style="background:transparent;" | ||
| − | |[[File: | + | |[[File:One_subject_acute_seg.png|323px|thumb|Segmentation of acute images of subject I]] |
| − | |[[File: | + | |[[File:One_subject_chronic_seg.png|323px|thumb|Segmentation of chronic images of subject I]] |
| − | |||
|- | |- | ||
|} | |} | ||
| + | </center> | ||
| + | |||
| + | = Former Work = | ||
| + | |||
| + | Whereas methodologies for segmentation of normal anatomical structures and tissue types has become standard and highly automated, tools for efficient and reliable segmentation of pathology lag behind. Major efforts on segmentation of lesions mostly focus on wm lesions as in multiple sclerosis and tumor/edema segmentation. | ||
| − | + | Driven by challenging image analysis problems of the NA-MIC DBP partner UCLA on [http://www.na-mic.org/Wiki/index.php/DBP3:UCLA#Introduction traumatic brain injury (TBI)], we will develop image segmentation methodology that helps clinical researcher to characterize and quantify a variety of different cerebral lesion types. Standard automated image analysis methods are not robust with respect to the TBI-related changes in image contrast, changes in brain shape, cranial fractures, lesions, white matter fiber alterations, and other signatures of head injury. | |
| + | |||
| + | We are working on an extension of the "atlas-based classification" method [http://www.na-mic.org/Wiki/index.php/Projects:UtahAtlasSegmentation ABC] for TBI datasets with the clinical goal to efficiently segment healthy brain tissue and cerebral lesions. A main goal will be the '''automated segmentation of healthy brain tissue''' and '''user-assisted segmentation of various cerebral lesion types''' (hematoma, subarachnoid hemorrhage, contusion and DAI, perifocal (regional) to diffuse (generalized) edema, hemorrhagic diffuse axonal injury (DAI)and more. | ||
| + | A strong emphasis will be on the joint analysis of multiple imaging modalities (T1 pre- and T1 postcontrast, T2 (TSE), FLAIR, GRE, SWI, Perfusion, and DTI/DWI) for improved detection and quantitative characterization of lesion types. | ||
| + | |||
| + | |||
| + | [[File:Multimodal_TBI.png|900px|center|thumb|Multimodal MRI data of TBI patient.]] | ||
| + | |||
| + | == Initial experiments == | ||
| − | = Experiments: Multi-modal registration and tissue segmentation = | + | === Experiments: Multi-modal registration and tissue segmentation === |
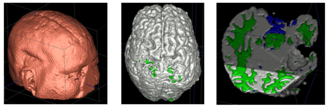
| − | We conducted experiments with the application of the ABC tool to multi-modal image data of 5 TBI cases provided by DBP partner UCLA. The tool includes co-registration of multiple modalities via mutual-information linear registration, and a nonlinear registration (high-deformable fluid registration) of a probabilistic normative atlas for segmentation of healthy tissue. The following results show feasibility of multi-modal registration and segmentation of normal tissue. Pathology is | + | We conducted experiments with the application of the ABC tool to multi-modal image data of 5 TBI cases provided by DBP partner UCLA. The tool includes co-registration of multiple modalities via mutual-information linear registration, and a nonlinear registration (high-deformable fluid registration) of a probabilistic normative atlas for segmentation of healthy tissue. The following results show feasibility of multi-modal registration and segmentation of normal tissue. Pathology is segmented via postprocessing using 3D user-supervised level-set evolution. |
| + | <center> | ||
{| border="0" style="background:transparent;" | {| border="0" style="background:transparent;" | ||
| − | |[[File:ABC-TBI-1.png| | + | |[[File:ABC-TBI-1.png|220px|thumb|Case 1: Segmentation of 5channel TBI data with ABC. Lesions and ventricles segmented with additional 3D level-set segmentation.]] |
| − | |[[File:ABC-TBI-5.png| | + | |[[File:ABC-TBI-5.png|310px|thumb|Case 2: Segmentation of 5channel TBI data with ABC. Lesions and ventricles segmented with additional 3D level-set segmentation.]] |
| − | |[[File:ABC-TBI-4.png| | + | |[[File:ABC-TBI-4.png|330px|thumb|Case 3: Segmentation of 5channel TBI data with ABC. Lesions and ventricles segmented with additional 3D level-set segmentation.]] |
|- | |- | ||
|} | |} | ||
| + | </center> | ||
Current work, jointly with the UCLA DBP partner, centers about a clinical definition of the most common lesion types and a multi-modality MRI characterization definition of these patterns. These characterizations will be used for the development of a segmentation methodology of the broad range of cerebral lesion types based on a user-guided definition of such patterns in current TBI cases. | Current work, jointly with the UCLA DBP partner, centers about a clinical definition of the most common lesion types and a multi-modality MRI characterization definition of these patterns. These characterizations will be used for the development of a segmentation methodology of the broad range of cerebral lesion types based on a user-guided definition of such patterns in current TBI cases. | ||
| − | = Experiments: Registration of longitudinal data = | + | === Experiments: Registration of longitudinal data === |
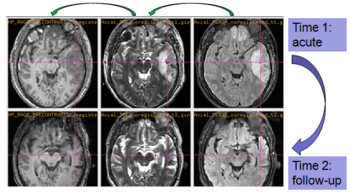
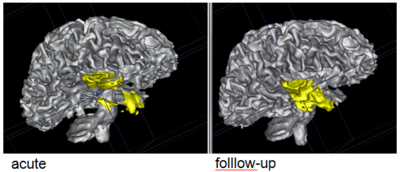
The NA-MIC DBP project on TBI analysis (UCLA partner) includes serial multi-modal MRI at acute phase and follow-up after six months. We test mutual-information-based linear registration of multi-modal MRI data within each time point and nonlinear registration (b-spline) of follow-up scans to obtain sets of images mapped into the same coordinate system. Preliminary results demonstrate the large deformations due to mass effect of a large lesion but also significant large regional changes of multi-modal MRI contrast from actute to follow-up. | The NA-MIC DBP project on TBI analysis (UCLA partner) includes serial multi-modal MRI at acute phase and follow-up after six months. We test mutual-information-based linear registration of multi-modal MRI data within each time point and nonlinear registration (b-spline) of follow-up scans to obtain sets of images mapped into the same coordinate system. Preliminary results demonstrate the large deformations due to mass effect of a large lesion but also significant large regional changes of multi-modal MRI contrast from actute to follow-up. | ||
| − | + | <center> | |
{| border="0" style="background:transparent;" | {| border="0" style="background:transparent;" | ||
| − | |[[File:TBI-long-reg.PNG| | + | |[[File:TBI-long-reg.PNG|350px|thumb|Longitudinal co-registration of multi-modal MRI.]] |
| − | |[[File:TBI-long-wmseg.PNG| | + | |[[File:TBI-long-wmseg.PNG|400px|thumb|Segmented white matter and temporal lobe (yellow) at acute phase and after 6 month follow-up.]] |
|- | |- | ||
|} | |} | ||
| + | </center> | ||
| + | |||
| + | === Conclusion === | ||
| + | |||
| + | In this work, we will develop new methodologies and tools for user-guided segmentation of cerebral lesions in multi-modal brain MRI. Driving application is TBI imaging research as conducted by the NA-MIC DBP partner UCLA. Components to be included will be robust clustering as presented in [8], initialization of multi-modal intensity patterns via prior knowledge [6], and evaluation of generative models as presented in [6] and [7]. Developments will also include efficient user-guidance for localization and characterization of subject-specific lesion patterns, and tools to provide quantitative measurements and indices of spatial extent of pathologies currently used in clinical assessment. We will evaluate extension of the commonly used indices derived from 2D images to 3D shape features. | ||
| + | |||
| + | = Collaboration events = | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | === (NA-MIC) Summer Project Week 2014 (June 23-27, 2014) === | ||
| + | Bo from Utah algorithm team remotely joined the summer project week and was working on understanding the workflow of Python scripting module, and modifying the existing code for a Slicer extension. Bo also worked on the documentation of the current command line tool for interactive TBI segmentation. | ||
| + | <br />[http://www.na-mic.org/Wiki/index.php/2014_Summer_Project_Week:TBI_Segmentation Webpage of this event] | ||
| + | |||
| + | === Software release to USC DBP Collaborators (February 13 - 14, 2014) === | ||
| + | |||
| + | Bo from Utah algorithm team went to visit USC TBO DBP team. Bo shared the interactive segmentation code of the command-line tool to the USC DBP team. Bo also did a presentation at the Slicer training workshop on February 14 to introduce the framework of the interactive segmentation for TBI patients. | ||
| + | <br />[http://www.na-mic.org/Wiki/index.php/USC_2014 Webpage of this event] | ||
| + | |||
| + | === (NA-MIC) All hands meeting (AHM) and Winter Project Week 2014 (January 6-10, 2014) === | ||
| + | |||
| + | During the project week, Bo from the Utah algorithm team ran the current interactive TBI segmentation command-line tool step by step with Andrei from USC DBP team, and showed how it worked. Utah team also had meeting with USC team to discuss sharing the current command line tool with them. We had a TBI DBP meeting as well to discuss with the Slicer community about the integration of current tool into Slicer. | ||
| + | <br />[http://www.na-mic.org/Wiki/index.php/2014_Project_Week:TBISegmentation Webpage of this event] | ||
| + | |||
| + | === (NA-MIC) Summer Project Week 2013 (June 17-21, 2013) === | ||
| + | |||
| + | Utah team coordinated with UCLA regarding the release of a Slicer module to perform multimodal automatic segmentation of TBI using the methodologies developed by the Utah team. Utah team also did a progress update to the UCLA group on the current status of the matlab/C++/VTK translation of the Utah algorithm. | ||
| + | <br />[http://www.na-mic.org/Wiki/index.php/Validation_and_testing_of_3D_Slicer_modules_implementing_the_Utah_segmentation_algorithm_for_traumatic_brain_injury Webpage of this event] | ||
| + | |||
| + | === Telephone meeting (April 1st, 2013) === | ||
| + | * Development of the multimodal registration and segmentation software. | ||
| + | * Journal article submission of current method. | ||
| + | * Grant reviews. | ||
| + | * Schedule of Andrei and Micah's visit. | ||
| + | |||
| + | === (NA-MIC) All hands meeting (AHM) and Winter Project Week 2013 (January 7-11) === | ||
| + | |||
| + | We explored the possibilities of combining structural and DTI information for assessment of recovery and treatment efficacy for TBI. We tried affine, Bspline, demons registration modules in Slicer to do coregistration of structural and DTI data of TBI. | ||
| + | <br />[http://www.na-mic.org/Wiki/index.php/4D_DTI_tractography_in_Slicer_for_TBI Webpage of this event] | ||
| + | |||
| + | === Telephone meeting (Sept. 17th, 2012) === | ||
| + | |||
| + | Andrei and Bo talked each other using skype, we discussed about the ongoing work of computation and visualization of cortical atrophy. | ||
| + | |||
| + | === (NA-MIC) Summer Project Week 2012 (July 18-22) === | ||
| + | We performed a systematic inspection of available UCLA TBI data. Problematic data sets were discussed and some additional data were obtained from UCLA for one data set. Bo and Andrei discussed to start working on two new TBI-related projects which may yield novel biomarkers of disease outcome. <br />[http://www.na-mic.org/Wiki/index.php/Semiautomatic_longitudinal_segmentation_of_MR_volumes_in_traumatic_brain_injury Webpage of this event] | ||
| + | |||
| + | === (NA-MIC) All hands meeting (AHM) and Winter Project Week 2012 (January 9-13) === | ||
| + | |||
| + | We got two new subjects from UCLA DBP. We processed the data and tested the current algorithm. We talked to our collaborators and learn the knowledge of pathology such as different lesions, necrosis and tissue compression. We also discussed the possible metrics we could provide to quantitatively analyze the tissue/lesion changes. <br />[http://www.na-mic.org/Wiki/index.php/2012_Winter_Project_Week:TBIClinicalAnalysis Webpage of this event] | ||
| + | === Telephone meeting (Sept. 9th, 2011) === | ||
| − | ==Key Investigators | + | During this telephone meeting, we discussed about the following problems. |
| + | * Integrate Mriwatcher to Slicer | ||
| + | * Manual segmentation and coregistration | ||
| + | * The SPIE submission | ||
| + | * The topological changes in TBI data | ||
| + | |||
| + | === (NA-MIC) Summer Project Week 2011 (July 20-24) === | ||
| + | |||
| + | We prepared the preliminary algorithm for this project week. During the project week, we applied the preliminary algorithm to current data and fixed some bugs in the code. Our collaborators helped us to validate our current results of the supervised segmentation. We got some comments and feedback from our collaborators, which are very important for us to improve the current algorithm. <br />[http://www.na-mic.org/Wiki/index.php/2011_Summer_Project_Week_Segmentation_TBI Webpage of this event] | ||
| + | |||
| + | === (NA-MIC) All hands meeting (AHM) and Winter Project Week 2011 (January 10-14) === | ||
| + | |||
| + | This is the first NA-MIC meeting between UCLA group and Utah group. During the project week, a lot of discussions have been done between the collaborators (UCLA and Utah groups) of this project. We worked together and answered each other's questions. Marcel introduced the purpose of all parameters of ABC and how these parameters would affect the final results. We discussed the possible user interaction we would use in the next step for better segmentation. Moreover, we knew more about the requirements of neurosurgeons and clinicians. Andrei pointed out the result of one subject was not good enough. Bo checked his results and redid the processing, and found the reason of the problem. <br />[http://www.na-mic.org/Wiki/index.php/2011_Winter_Project_Week:Extension_of_ABC_to_detect_pathology_categories Webpage of this event] | ||
| + | |||
| + | = Literature = | ||
| + | |||
| + | [1] Bo Wang, Wei Liu, Marcel Prastawa, Andrei Irimia, Paul M. Vespa, John D. Van Horn, P. Thomas Fletcher, and Guido Gerig . 4D Active Cut: An Interactive Tool for Pathological Anatomy Modeling, In Biomedical Imaging (ISBI), 2014 IEEE 11th International Symposium on, pp. 529-532. IEEE, 2014. | ||
| + | |||
| + | [2] BoWang, Marcel Prastawa, Avishek Saha, Suyash P. Awate, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. Van Horn, Valerio Pascucci, and Guido Gerig., Modeling 4D Changes in Pathological Anatomy Using Domain Adaptation: Analysis of TBI Imaging Using a Tumor Database, In Multimodal Brain Image Analysis, pp. 31-39. 2013. [Best Paper Award] | ||
| + | |||
| + | [3] Bo Wang, Marcel Prastawa, Andrei Irimia, Micah C. Chambers, Neda Sadeghi, Paul M. Vespa, John D. Van Horn, Guido Gerig, Analyzing Imaging Biomarkers for Traumatic Brain Injury Using 4D Modeling of Longitudinal MRI, In IEEE Proceedings of ISBI 2013, pp. 1392-1395. 2013. | ||
| + | |||
| + | [4] Bo Wang, Marcel Prastawa, Suyash P. Awate, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. Van Horn, Guido Gerig, Segmentation of Serial MRI of TBI patients using Personalized Atlas Construction and Topological Change Estimation, In Proceedings of IEEE ISBI 2012, pp. 1152--1155. 2012. | ||
| + | |||
| + | [5] Bo Wang, Marcel Prastawa, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. Van Horn, and Guido Gerig, A Patient-Specific Segmentation Framework for Longitudinal MR Images of Traumatic Brain Injury, Proc. SPIE 8314, 831402 (2012). | ||
| + | |||
| + | [6] Marcel Prastawa, Elizabeth Bullitt, Guido Gerig, Simulation of brain tumors in MR images, Medical Image Analysis 13 (2009), pp. 297-311, PMID: 19119055 | ||
| + | |||
| + | [7] Marcel Prastawa and Guido Gerig, Brain Lesion Segmentation through Physical Model Estimation, International Symposium on Visual Computing ISVC 2008, LNCS, Vol. 5358, pp. 562--571. | ||
| + | |||
| + | [8] Marcel Prastawa, John H. Gilmore, Weili Lin, Guido Gerig, Automatic Segmentation of MR Images of the Developing Newborn Brain, Medical Image Analysis (MedIA). Vol 9, October 2005, pages 457-466 | ||
| + | |||
| + | =Key Investigators= | ||
* Utah: Bo Wang, Marcel Prastawa, Guido Gerig | * Utah: Bo Wang, Marcel Prastawa, Guido Gerig | ||
| − | * UCLA: | + | * UCLA: John D. Van Horn, Andrei Irimia, Micah Chambers / Clinical partners: Paul Vespa, M.D., David Hovda, M.D. |
| + | |||
| − | + | =References= | |
* [http://www.na-mic.org/Wiki/index.php/Projects:UtahAtlasSegmentation ABC NA-MIC Project] | * [http://www.na-mic.org/Wiki/index.php/Projects:UtahAtlasSegmentation ABC NA-MIC Project] | ||
* [http://www.nitrc.org/projects/abc ABC nitrc] | * [http://www.nitrc.org/projects/abc ABC nitrc] | ||
* [http://www.na-mic.org/Wiki/index.php/DBP3:UCLA Traumatic brain injury (TBI)] | * [http://www.na-mic.org/Wiki/index.php/DBP3:UCLA Traumatic brain injury (TBI)] | ||
| + | |||
| + | <br /> | ||
| + | Back to [[Algorithm:Utah2|Utah 2 Algorithms]] | ||
Latest revision as of 23:55, 11 October 2014
Home < Projects:PathologyAnalysisBack to Utah 2 Algorithms
Ongoing Work
4D Active Cut: An Interactive Tool for Pathological Anatomy Modeling
We propose a novel semi-supervised method, called 4D active cut for lesion recognition and deformation estimation. Existing interactive segmentation methods passively wait for user to refine the segmentations which is a difficult task in 3D images that change over time. 4D active cut instead actively selects candidate regions for querying the user, and therefore obtains the most informative user feedback. A user simply answers `yes' or `no' to a candidate object without having to refine the segmentation slice by slice. Compared to single-object detection of GrabCut, our method also detects multiple lesions with spatial coherence using Markov random fields constraints. [1]
The following figure shows the dynamic learning process of the proposed framework applied to one subject.
Modeling 4D Changes in Pathological Anatomy
We propose a novel framework that models 4D changes in pathological anatomy across time, and provides explicit mapping from a healthy template to subjects with pathology. Moreover, our framework uses transfer learning to leverage rich information from a known source domain, where we have a collection of completely segmented images, to yield effective appearance models for the input target domain. The automatic 4D segmentation method uses a novel domain adaptation technique for generative kernel density models to transfer information between different domains, resulting in a fully automatic method that requires no user interaction. [2]
Analyzing imaging biomarkers for traumatic brain injury using 4D modeling of longitudinal MRI
We present a new method for computing surface-based and voxel-based imaging biomarkers using 4D modeling of longitudinal MRI. We analyze the potential for clinical use of these biomarkers by correlating them with TBI-specific patient scores. Our preliminary results indicate that the proposed voxel-based biomarkers are correlated with clinical outcomes [3].
Segmentation of Serial MRI of TBI patients using Personalized Atlas Construction and Topological Change Estimation
Longitudinal images with TBI present topological changes over time due to the effect of the impact force on tissue, skull, and blood vessels and the recovery process. We address this issue by defining a novel atlas construction scheme that explicitly models the effect of topological changes [4]. Our method automatically estimates the probability of topological changes jointly with the personalized atlas. The following figure shows our atlas construction scheme.
Our method combines 4D information through the creation of personalized atlas that explicitly handles diffeomorphic and nondiffeomorphic temporal changes. The method is robust to topological changes caused by the injury and the recovery process in TBI.
Patient-Specific Segmentation Framework for Longitudinal MR Images of Traumatic Brain Injury
We propose a multi-modal image segmentation framework for longitudinal TBI images [5]. The framework is initialized through manual input of primary lesion sites at each time point, which are then refined by a joint approach composed of Bayesian segmentation and construction of a personalized atlas. The personalized atlas construction estimates the average of the posteriors of the Bayesian segmentation at each time point and warps the average back to each time point to provide the updated priors for Bayesian segmentation. The difference between our approach and segmenting longitudinal images independently is that we use the information from all time points to improve the segmentations. Given a manual initialization, our framework automatically segments healthy structures (white matter, grey matter, cerebrospinal fluid) as well as different lesions such as hemorrhagic lesions and edema. Our framework can handle different sets of modalities at each time point, which provides flexibility in analyzing clinical scans.
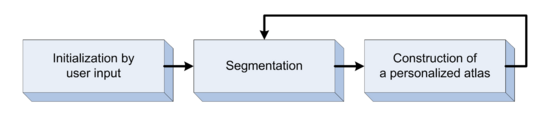
Segmentation framework
The segmentation framework is initialized through manual input of primary lesion sites at each time point, which are then refined by a joint approach composed of Bayesian segmentation and construction of a personalized atlas. The following figure is the segmentation framework for multi-modal MR images.
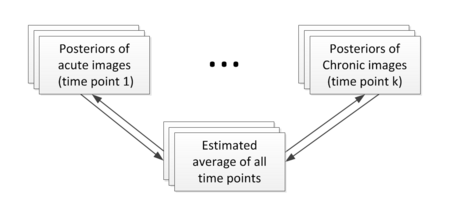
Construction of personalized atlas
Based on the segmentation results (the posteriors), we create personalized atlases using the unbiased diffeomorphic atlas construction method. In atlas construction, we estimate an average that requires the minimum amount of deformation to transform into the posteriors at every time point. The following flowchart shows the estimation of personalized atlases.
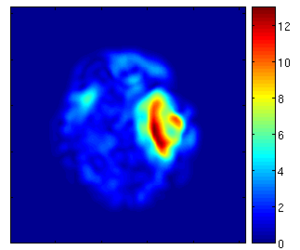
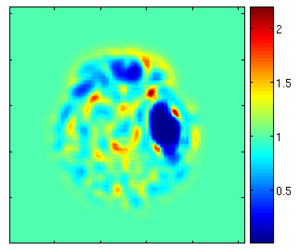
Results
Using the deformation field got from the estimation of personalized atlases, we can visualize the tissue deformation. In the following two figures, the deformation field is visualized as vector magnitude (left) and Jacobian (right).
The following two figures show the snapshot of the segmentation result.
Former Work
Whereas methodologies for segmentation of normal anatomical structures and tissue types has become standard and highly automated, tools for efficient and reliable segmentation of pathology lag behind. Major efforts on segmentation of lesions mostly focus on wm lesions as in multiple sclerosis and tumor/edema segmentation.
Driven by challenging image analysis problems of the NA-MIC DBP partner UCLA on traumatic brain injury (TBI), we will develop image segmentation methodology that helps clinical researcher to characterize and quantify a variety of different cerebral lesion types. Standard automated image analysis methods are not robust with respect to the TBI-related changes in image contrast, changes in brain shape, cranial fractures, lesions, white matter fiber alterations, and other signatures of head injury.
We are working on an extension of the "atlas-based classification" method ABC for TBI datasets with the clinical goal to efficiently segment healthy brain tissue and cerebral lesions. A main goal will be the automated segmentation of healthy brain tissue and user-assisted segmentation of various cerebral lesion types (hematoma, subarachnoid hemorrhage, contusion and DAI, perifocal (regional) to diffuse (generalized) edema, hemorrhagic diffuse axonal injury (DAI)and more. A strong emphasis will be on the joint analysis of multiple imaging modalities (T1 pre- and T1 postcontrast, T2 (TSE), FLAIR, GRE, SWI, Perfusion, and DTI/DWI) for improved detection and quantitative characterization of lesion types.
Initial experiments
Experiments: Multi-modal registration and tissue segmentation
We conducted experiments with the application of the ABC tool to multi-modal image data of 5 TBI cases provided by DBP partner UCLA. The tool includes co-registration of multiple modalities via mutual-information linear registration, and a nonlinear registration (high-deformable fluid registration) of a probabilistic normative atlas for segmentation of healthy tissue. The following results show feasibility of multi-modal registration and segmentation of normal tissue. Pathology is segmented via postprocessing using 3D user-supervised level-set evolution.
Current work, jointly with the UCLA DBP partner, centers about a clinical definition of the most common lesion types and a multi-modality MRI characterization definition of these patterns. These characterizations will be used for the development of a segmentation methodology of the broad range of cerebral lesion types based on a user-guided definition of such patterns in current TBI cases.
Experiments: Registration of longitudinal data
The NA-MIC DBP project on TBI analysis (UCLA partner) includes serial multi-modal MRI at acute phase and follow-up after six months. We test mutual-information-based linear registration of multi-modal MRI data within each time point and nonlinear registration (b-spline) of follow-up scans to obtain sets of images mapped into the same coordinate system. Preliminary results demonstrate the large deformations due to mass effect of a large lesion but also significant large regional changes of multi-modal MRI contrast from actute to follow-up.
Conclusion
In this work, we will develop new methodologies and tools for user-guided segmentation of cerebral lesions in multi-modal brain MRI. Driving application is TBI imaging research as conducted by the NA-MIC DBP partner UCLA. Components to be included will be robust clustering as presented in [8], initialization of multi-modal intensity patterns via prior knowledge [6], and evaluation of generative models as presented in [6] and [7]. Developments will also include efficient user-guidance for localization and characterization of subject-specific lesion patterns, and tools to provide quantitative measurements and indices of spatial extent of pathologies currently used in clinical assessment. We will evaluate extension of the commonly used indices derived from 2D images to 3D shape features.
Collaboration events
(NA-MIC) Summer Project Week 2014 (June 23-27, 2014)
Bo from Utah algorithm team remotely joined the summer project week and was working on understanding the workflow of Python scripting module, and modifying the existing code for a Slicer extension. Bo also worked on the documentation of the current command line tool for interactive TBI segmentation.
Webpage of this event
Software release to USC DBP Collaborators (February 13 - 14, 2014)
Bo from Utah algorithm team went to visit USC TBO DBP team. Bo shared the interactive segmentation code of the command-line tool to the USC DBP team. Bo also did a presentation at the Slicer training workshop on February 14 to introduce the framework of the interactive segmentation for TBI patients.
Webpage of this event
(NA-MIC) All hands meeting (AHM) and Winter Project Week 2014 (January 6-10, 2014)
During the project week, Bo from the Utah algorithm team ran the current interactive TBI segmentation command-line tool step by step with Andrei from USC DBP team, and showed how it worked. Utah team also had meeting with USC team to discuss sharing the current command line tool with them. We had a TBI DBP meeting as well to discuss with the Slicer community about the integration of current tool into Slicer.
Webpage of this event
(NA-MIC) Summer Project Week 2013 (June 17-21, 2013)
Utah team coordinated with UCLA regarding the release of a Slicer module to perform multimodal automatic segmentation of TBI using the methodologies developed by the Utah team. Utah team also did a progress update to the UCLA group on the current status of the matlab/C++/VTK translation of the Utah algorithm.
Webpage of this event
Telephone meeting (April 1st, 2013)
- Development of the multimodal registration and segmentation software.
- Journal article submission of current method.
- Grant reviews.
- Schedule of Andrei and Micah's visit.
(NA-MIC) All hands meeting (AHM) and Winter Project Week 2013 (January 7-11)
We explored the possibilities of combining structural and DTI information for assessment of recovery and treatment efficacy for TBI. We tried affine, Bspline, demons registration modules in Slicer to do coregistration of structural and DTI data of TBI.
Webpage of this event
Telephone meeting (Sept. 17th, 2012)
Andrei and Bo talked each other using skype, we discussed about the ongoing work of computation and visualization of cortical atrophy.
(NA-MIC) Summer Project Week 2012 (July 18-22)
We performed a systematic inspection of available UCLA TBI data. Problematic data sets were discussed and some additional data were obtained from UCLA for one data set. Bo and Andrei discussed to start working on two new TBI-related projects which may yield novel biomarkers of disease outcome.
Webpage of this event
(NA-MIC) All hands meeting (AHM) and Winter Project Week 2012 (January 9-13)
We got two new subjects from UCLA DBP. We processed the data and tested the current algorithm. We talked to our collaborators and learn the knowledge of pathology such as different lesions, necrosis and tissue compression. We also discussed the possible metrics we could provide to quantitatively analyze the tissue/lesion changes.
Webpage of this event
Telephone meeting (Sept. 9th, 2011)
During this telephone meeting, we discussed about the following problems.
- Integrate Mriwatcher to Slicer
- Manual segmentation and coregistration
- The SPIE submission
- The topological changes in TBI data
(NA-MIC) Summer Project Week 2011 (July 20-24)
We prepared the preliminary algorithm for this project week. During the project week, we applied the preliminary algorithm to current data and fixed some bugs in the code. Our collaborators helped us to validate our current results of the supervised segmentation. We got some comments and feedback from our collaborators, which are very important for us to improve the current algorithm.
Webpage of this event
(NA-MIC) All hands meeting (AHM) and Winter Project Week 2011 (January 10-14)
This is the first NA-MIC meeting between UCLA group and Utah group. During the project week, a lot of discussions have been done between the collaborators (UCLA and Utah groups) of this project. We worked together and answered each other's questions. Marcel introduced the purpose of all parameters of ABC and how these parameters would affect the final results. We discussed the possible user interaction we would use in the next step for better segmentation. Moreover, we knew more about the requirements of neurosurgeons and clinicians. Andrei pointed out the result of one subject was not good enough. Bo checked his results and redid the processing, and found the reason of the problem.
Webpage of this event
Literature
[1] Bo Wang, Wei Liu, Marcel Prastawa, Andrei Irimia, Paul M. Vespa, John D. Van Horn, P. Thomas Fletcher, and Guido Gerig . 4D Active Cut: An Interactive Tool for Pathological Anatomy Modeling, In Biomedical Imaging (ISBI), 2014 IEEE 11th International Symposium on, pp. 529-532. IEEE, 2014.
[2] BoWang, Marcel Prastawa, Avishek Saha, Suyash P. Awate, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. Van Horn, Valerio Pascucci, and Guido Gerig., Modeling 4D Changes in Pathological Anatomy Using Domain Adaptation: Analysis of TBI Imaging Using a Tumor Database, In Multimodal Brain Image Analysis, pp. 31-39. 2013. [Best Paper Award]
[3] Bo Wang, Marcel Prastawa, Andrei Irimia, Micah C. Chambers, Neda Sadeghi, Paul M. Vespa, John D. Van Horn, Guido Gerig, Analyzing Imaging Biomarkers for Traumatic Brain Injury Using 4D Modeling of Longitudinal MRI, In IEEE Proceedings of ISBI 2013, pp. 1392-1395. 2013.
[4] Bo Wang, Marcel Prastawa, Suyash P. Awate, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. Van Horn, Guido Gerig, Segmentation of Serial MRI of TBI patients using Personalized Atlas Construction and Topological Change Estimation, In Proceedings of IEEE ISBI 2012, pp. 1152--1155. 2012.
[5] Bo Wang, Marcel Prastawa, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. Van Horn, and Guido Gerig, A Patient-Specific Segmentation Framework for Longitudinal MR Images of Traumatic Brain Injury, Proc. SPIE 8314, 831402 (2012).
[6] Marcel Prastawa, Elizabeth Bullitt, Guido Gerig, Simulation of brain tumors in MR images, Medical Image Analysis 13 (2009), pp. 297-311, PMID: 19119055
[7] Marcel Prastawa and Guido Gerig, Brain Lesion Segmentation through Physical Model Estimation, International Symposium on Visual Computing ISVC 2008, LNCS, Vol. 5358, pp. 562--571.
[8] Marcel Prastawa, John H. Gilmore, Weili Lin, Guido Gerig, Automatic Segmentation of MR Images of the Developing Newborn Brain, Medical Image Analysis (MedIA). Vol 9, October 2005, pages 457-466
Key Investigators
- Utah: Bo Wang, Marcel Prastawa, Guido Gerig
- UCLA: John D. Van Horn, Andrei Irimia, Micah Chambers / Clinical partners: Paul Vespa, M.D., David Hovda, M.D.
References
Back to Utah 2 Algorithms