Difference between revisions of "Projects:PathologyAnalysis"
| Line 9: | Line 9: | ||
On anatomical MRI scans, to quantitatively analyze the cortical thickness, white matter changes, we need to have a good segmentation on TBI images. However, for TBI data, standard automated image analysis methods are not robust with respect to the TBI-related changes in image contrast, changes in brain shape, cranial fractures, white matter fiber alterations, and other signatures of head injury. | On anatomical MRI scans, to quantitatively analyze the cortical thickness, white matter changes, we need to have a good segmentation on TBI images. However, for TBI data, standard automated image analysis methods are not robust with respect to the TBI-related changes in image contrast, changes in brain shape, cranial fractures, white matter fiber alterations, and other signatures of head injury. | ||
| − | |||
| − | |||
| − | |||
{| border="0" style="background:transparent;" | {| border="0" style="background:transparent;" | ||
| Line 24: | Line 21: | ||
|} | |} | ||
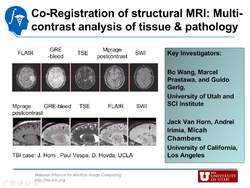
| − | + | We are working on an extension of the "atlas-based classification" method ABC [http://www.na-mic.org/Wiki/index.php/Projects:UtahAtlasSegmentation] for TBI datasets with the clinical goal to efficiently segment healthy brain tissue and cerebral lesions. A main goal will be the '''automated segmentation healthy brain tissue''' and '''user-assisted segmentation of various cerebral lesion types''' (hematoma, subarachnoid hemorrhage, contusion and DAI, perifocal (regional) to diffuse (generalized) edema, hemorrhagic diffuse axonal injury (DAI)and more. | |
| + | A strong emphasis will be on the joint of multiple imaging modalities (T1 pre- and T1 postcontrast, T2 (TSE), FLAIR, GRE, SWI, Perfusion, and DTI/DWI) for improved detection and quantitative characterization of lesion types. | ||
==Key Investigators== | ==Key Investigators== | ||
Revision as of 17:09, 11 April 2011
Home < Projects:PathologyAnalysisBack to Utah 2 Algorithms
Analysis of Brain Images with Variety of Cerebral Lesion Types
Description
Traumatic brain injury (TBI) occurs when an external force traumatically injures the brain. TBI is a major cause of death and disability worldwide, especially in children and young adults. TBI affects 1.4 million Americans annually. The UCLA medical school has been working on this topic for years.
On anatomical MRI scans, to quantitatively analyze the cortical thickness, white matter changes, we need to have a good segmentation on TBI images. However, for TBI data, standard automated image analysis methods are not robust with respect to the TBI-related changes in image contrast, changes in brain shape, cranial fractures, white matter fiber alterations, and other signatures of head injury.
We are working on an extension of the "atlas-based classification" method ABC [1] for TBI datasets with the clinical goal to efficiently segment healthy brain tissue and cerebral lesions. A main goal will be the automated segmentation healthy brain tissue and user-assisted segmentation of various cerebral lesion types (hematoma, subarachnoid hemorrhage, contusion and DAI, perifocal (regional) to diffuse (generalized) edema, hemorrhagic diffuse axonal injury (DAI)and more. A strong emphasis will be on the joint of multiple imaging modalities (T1 pre- and T1 postcontrast, T2 (TSE), FLAIR, GRE, SWI, Perfusion, and DTI/DWI) for improved detection and quantitative characterization of lesion types.
Key Investigators
- Utah: Bo Wang, Marcel Prastawa, Guido Gerig
- UCLA: Jack Van Horn, Andrei Irimia, Micah Chambers