Projects:PathologyAnalysis
Back to Utah 2 Algorithms
Analysis of Brain Images with Variety of Cerebral Lesion Types
Description
Traumatic brain injury (TBI) occurs when an external force traumatically injures the brain. TBI is a major cause of death and disability worldwide, especially in children and young adults. TBI affects 1.4 million Americans annually. The UCLA medical school has been working on this topic for years.
On anatomical MRI scans, to quantitatively analyze the cortical thickness, white matter changes, we need to have a good segmentation on TBI images. However, for TBI data, standard automated image analysis methods are not robust with respect to the TBI-related changes in image contrast, changes in brain shape, cranial fractures, white matter fiber alterations, and other signatures of head injury.
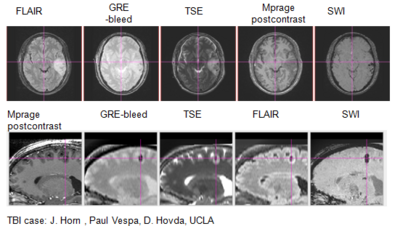
We are working on an extension of the "atlas-based classification" method ABC [1] for TBI datasets with the clinical goal to efficiently segment healthy brain tissue and cerebral lesions. A main goal will be the automated segmentation of healthy brain tissue and user-assisted segmentation of various cerebral lesion types (hematoma, subarachnoid hemorrhage, contusion and DAI, perifocal (regional) to diffuse (generalized) edema, hemorrhagic diffuse axonal injury (DAI)and more. A strong emphasis will be on the joint of multiple imaging modalities (T1 pre- and T1 postcontrast, T2 (TSE), FLAIR, GRE, SWI, Perfusion, and DTI/DWI) for improved detection and quantitative characterization of lesion types.
Experiments: Multi-modal registration and tissue segmentation
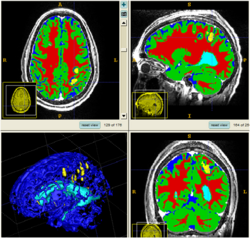
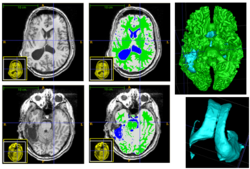
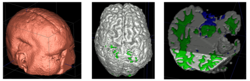
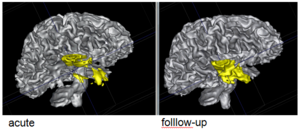
We conducted experiments with the application of the ABC tool to multi-modal image data of 5 TBI cases provided by DBP partner UCLA. The tool includes co-registration of multiple modalities via mutual-information linear registration, and a nonlinear registration (high-deformable fluid registration) of a probabilistic normative atlas for segmentation of healthy tissue. The following results show feasibility of multi-modal registration and segmentation of normal tissue. Pathology is currently segmented via postprocessing using 3D user-supervised level-set evolution.
Current work, jointly with the UCLA DBP partner, centers about a clinical definition of the most common lesion types and a multi-modality MRI characterization definition of these patterns. These characterizations will be used for the development of a segmentation methodology of the broad range of cerebral lesion types based on a user-guided definition of such patterns in current TBI cases.
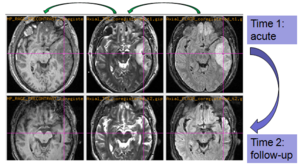
Experiments: Registration of longitudinal data
The NA-MIC DBP project on TBI analysis (UCLA partner) includes serial multi-modal MRI at acute phase and follow-up after six months. We test mutual-information-based linear registration of multi-modal MRI data within each time point and nonlinear registration (b-spline) of follow-up scans to obtain sets of images mapped into the same coordinate system. Preliminary results demonstrate the large deformations due to mass effect of a large lesion but also significant large regional changes of multi-modal MRI contrast from actute to follow-up.
Conclusion
In this work, we will develop new methodologies and tools for user-guided segmentation of cerebral lesions in multi-modal brain MRI. Driving application is TBI imaging research as conducted by the NA-MIC DBP partner UCLA. Components to be included will be robust clustering as presented in [3], initialization of multi-modal intensity patterns via prior knowledge [3], and evaluation of generative models as presented in [1] and [2]. Developments will also include efficient user-guidance for localization and characterization of subject-specific lesion patterns, and tools to provide quantitative measurements and indices of spatial extent of pathologies currently used in clinical assessment. We will evaluate extension of the commonly used indices derived from 2D images to 3D shape features.
Literature
[1] Prastawa M, Bullitt E, Gerig G, Simulation of brain tumors in MR images, Medical Image Analysis 13 (2009), pp. 297-311, PMID: 19119055 [2] Marcel Prastawa and Guido Gerig, Brain Lesion Segmentation through Physical Model Estimation, In Lecture Notes in Computer Science, Vol. 5358, pp. 562--571. 2008. [3] Marcel Prastawa, John H. Gilmore, Weili Lin, Guido Gerig, Automatic Segmentation of MR Images of the Developing Newborn Brain, Medical Image Analysis (MedIA). Vol 9, October 2005, pages 457-466
Key Investigators
- Utah: Bo Wang, Marcel Prastawa, Guido Gerig
- UCLA: Jack Van Horn, Andrei Irimia, Micah Chambers / Clinical partners: Paul Vespa, M.D., David Hovda, M.D.