Difference between revisions of "Projects:RegistrationLibrary:RegLib C05"
From NAMIC Wiki
(→Input) |
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
||
| Line 17: | Line 17: | ||
=== Modules === | === Modules === | ||
| − | *'''Slicer 3.6.1 recommended modules: [ | + | *'''Slicer 3.6.1 recommended modules: [https://www.slicer.org/wiki/Modules:PythonSurfaceICPRegistration-Documentation-3.6 Surface Registration] |
===Objective / Background === | ===Objective / Background === | ||
Latest revision as of 17:06, 10 July 2017
Home < Projects:RegistrationLibrary:RegLib C05Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.1  Slicer Registration Library Case 05:
Slicer Registration Library Case 05:
Inter-subject Knee MRI
Input
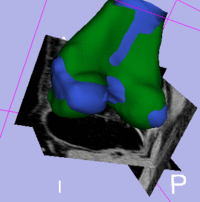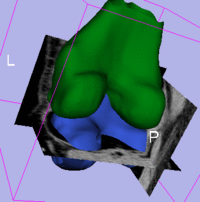
|
|
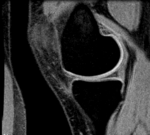
|
| subject 1 | subject 1 |
Modules
- Slicer 3.6.1 recommended modules: Surface Registration
Objective / Background
The final goal is to align a segmentation prior model/atlas to aid in cartilage segmentation.
Keywords
MRI, knee, inter-subject, segmentation
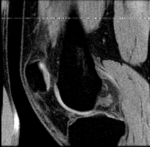
Input Data
- fixed: T1 SPGR , 0.9375 x 0.9375 x 1.4 mm voxel size, axial. + surface model from femur & tibia segmentation.
- moving: T1 SPGR , 0.9375 x 0.9375 x 1.2 mm voxel size, sagittal. + surface model from femur & tibia segmentation.
Procedure / Pipeline
- Open case scene file or import image data + surface models
- Open Registration / Surface Registration module
- Input Surface: S02_femur
- Target Surface: S01_femur
- Output Transform: create new transform, rename to "Xf1_surf_S2-S1"
- Select/check Mean distance mode: RMS
- Leave defaults: iterations: 50, landmarks: 200
- Select/check Start by matching centroids
- Select/check Check mean distance
- Click: Apply; process duration: ~ seconds
- Go to the Data module
- find the newly created transform node "Xf1_surf_S2-S1" and drag the "S02_femur" model node (and all other S02 entities) inside the transform.
- for the 3D viewer turn on the visibility of the surface models (should be on already)
- the surfaces for the two femur models should now align in basic position and orientation
- for additional details see tutorial downloads below
Registration Results
Download
- Data
- Presets
- Link to User Guide: How to Load/Save Registration Parameter Presets
- Documentation