Difference between revisions of "Projects:RegistrationLibrary:RegLib C06"
From NAMIC Wiki
| Line 76: | Line 76: | ||
##''Apply'' | ##''Apply'' | ||
#repeat for Post_Rx_left_n4 | #repeat for Post_Rx_left_n4 | ||
| + | #save intermediate results | ||
| + | #we need to exclude the tumor from the mask, to prevent the registration from trying to match that region. Much should already be excluded from the above segmentation, but it may be necessary to apply some manual edits: | ||
| + | #Go to the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/Editor ''Editor''] module | ||
| + | ##Click ''Apply'' on the popup do confirm colormap selection (choice does not matter) | ||
| + | ##Master Volume: PreRx_left | ||
| + | ##''Merge Volume'': PreRx_mask (generated above) | ||
| + | ##click on the Brush Icon ("PaintEffect") | ||
| + | ##Label: 0 (black) | ||
| + | ##increase radius to ~ 10-15 mm. | ||
| + | ##navigate to an axial slice showing the tumor | ||
| + | ##click & drag left mouse to apply, then use arrow keys to move to the next slice. | ||
| + | #repeat for Post_Rx_mask | ||
#save intermediate results | #save intermediate results | ||
*'''Phase 4: Affine pre-Registration''' | *'''Phase 4: Affine pre-Registration''' | ||
| Line 90: | Line 102: | ||
##Leave all other settings at default | ##Leave all other settings at default | ||
##''Apply'' | ##''Apply'' | ||
| − | ##this should generate a first alignment. | + | ##this should generate a first alignment. note that the Slicer4.1 version of BRAINS registration does '''not''' automatically place the moving volume inside the result transform, as it did for Slicer 3.6. In order to see the result, you must either go to the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/Data ''Data module''] and drag the moving volume inside the transform node, or select to generate a resampled ''Output Volume''. See demo movie you can download here shows how to do this. |
| − | + | *'''Phase 5: Nonrigid final registration''' | |
| − | + | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit ''General Registration (BRAINS)''] module | |
| − | + | ##''Fixed Image Volume'': PreRx_left_n4 | |
| − | + | ##''Moving Image Volume'': PostRx_left_n4 | |
| − | |||
| − | |||
| − | |||
| − | *'''Phase | ||
| − | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module | ||
| − | ##''Fixed Image Volume'': | ||
| − | ##''Moving Image Volume'': | ||
##Output Settings: | ##Output Settings: | ||
| − | ###''Slicer BSpline Transform": create new transform, rename to " | + | ###''Slicer BSpline Transform": create & rename new transform, rename to "Xf2_BSpline" |
###''Slicer Linear Transform'': none | ###''Slicer Linear Transform'': none | ||
| − | ###''Output Image Volume'': create new | + | ###''Output Image Volume'': create & rename new transform, rename to "PostRx_left_Xf2" |
| − | ##''Registration Phases'': check | + | ##''Registration Phases'': uncheck rigid & affine boxes, check box for ''BSpline'' only |
##''Main Parameters'': | ##''Main Parameters'': | ||
| − | ###''Number Of Samples'': | + | ###''Number Of Samples'': 300,000 |
| − | ###''B-Spline Grid Size'': | + | ###''B-Spline Grid Size'': 7,7,5 |
##''Mask Option'': select ''ROI'' button | ##''Mask Option'': select ''ROI'' button | ||
| − | ###''ROI Masking input fixed'': select " " | + | ###''ROI Masking input fixed'': select "PreRx_mask" generated in Phase 3 above |
| − | ###''ROI Masking input moving'': select " | + | ###''ROI Masking input moving'': select "PostRx_mask" generated in Phase 3 above |
##Leave all other settings at default | ##Leave all other settings at default | ||
##click: Apply | ##click: Apply | ||
Revision as of 22:21, 5 June 2012
Home < Projects:RegistrationLibrary:RegLib C06Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
updated for v4.1  Registration Library Case #6: Breast MRI Treatment Assessment
Registration Library Case #6: Breast MRI Treatment Assessment
|
|
|
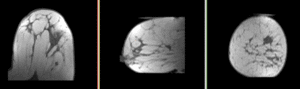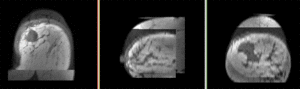
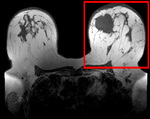
| fixed image/target pre Rx MRI |
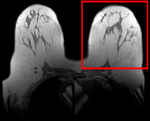
moving image post Rx MRI |
Versions
For the Slicer 3.6 version of this tutorial see here
Modules
- Slicer 3.6.1 recommended modules:
Objective / Background
We seek to align the post-treatment (PostRx) scan with the pre-treatment scan to compare local effects (left side only).
Keywords
MRI, breast cancer, intra-subject, treatment assessment, change detection, non-rigid registration
Download
- Data:
- Presets
- Documentation
Input Data
- reference/fixed : 0.44 x 0.44 x 5 mm , 784 x 784 x 30
- moving: 0.68 x 0.68 x 1.5 mm, 515 x 515 x 93
Methods
- Phase 1: Extract left breast image of PreRx and PostRx scan
- Open the Crop Volume module
- Input volume: "PreRx"
- Input ROI: "Create new annotation ROI"
- ROI visibilité: turn on
- Isotropic output voxel: yes (checkbox)
- Interpolator: "WindowedSinc" (radio button)
- place/drag the color markers visible in the slice views to enclose the left breast only (right side of image)
- click on Crop! button
- go to the Data module
- several new nodes were created: look for the "PreRx_subvolume-scale_1" entry, double click and rename to "PreRx_Left"
- repeat the same for the "PostRx" image
- Save intermediate results.
- Phase 2: MRI Bias field inhomogeneity correction
- Open the N4ITKBiasFieldCorrection module
- Input Image: PreRx_left
- Mask Image: none
- Output Volume: create & rename new: "PreRx_left_n4"
- leave all parameters at defaults
- Apply.
- repeat for the Post_Rx image.
- save intermediate results
- Phase 3: Build masks
- open the Foregroud masking (BRAINS) module (under Segmentation:Specialized)
- Input Image Volume: PreRx_left
- Output Mask: create & rename new: "PreRx_mask"
- Output Image clipped by ROI: none
- Configuration Parameters:
- ROI Auto Dilate Size: 0.1
- defaults for the rest
- Apply
- repeat for Post_Rx_left_n4
- save intermediate results
- we need to exclude the tumor from the mask, to prevent the registration from trying to match that region. Much should already be excluded from the above segmentation, but it may be necessary to apply some manual edits:
- Go to the Editor module
- Click Apply on the popup do confirm colormap selection (choice does not matter)
- Master Volume: PreRx_left
- Merge Volume: PreRx_mask (generated above)
- click on the Brush Icon ("PaintEffect")
- Label: 0 (black)
- increase radius to ~ 10-15 mm.
- navigate to an axial slice showing the tumor
- click & drag left mouse to apply, then use arrow keys to move to the next slice.
- repeat for Post_Rx_mask
- save intermediate results
- Phase 4: Affine pre-Registration
- open the General Registration (BRAINS) module
- Fixed Image Volume: PreRx_left_n4
- Moving Image Volume: PostRx_left_n4
- Output Settings:
- Slicer BSpline Transform": none
- Slicer Linear Transform: create & rename new transform, rename to "Xf1_Affine"
- Output Image Volume: none
- Registration Phases: check boxes for Rigid , Rigid+Scale and Affine
- Main Parameters:
- Number Of Samples: 200,000
- Leave all other settings at default
- Apply
- this should generate a first alignment. note that the Slicer4.1 version of BRAINS registration does not automatically place the moving volume inside the result transform, as it did for Slicer 3.6. In order to see the result, you must either go to the Data module and drag the moving volume inside the transform node, or select to generate a resampled Output Volume. See demo movie you can download here shows how to do this.
- Phase 5: Nonrigid final registration
- open the General Registration (BRAINS) module
- Fixed Image Volume: PreRx_left_n4
- Moving Image Volume: PostRx_left_n4
- Output Settings:
- Slicer BSpline Transform": create & rename new transform, rename to "Xf2_BSpline"
- Slicer Linear Transform: none
- Output Image Volume: create & rename new transform, rename to "PostRx_left_Xf2"
- Registration Phases: uncheck rigid & affine boxes, check box for BSpline only
- Main Parameters:
- Number Of Samples: 300,000
- B-Spline Grid Size: 7,7,5
- Mask Option: select ROI button
- ROI Masking input fixed: select "PreRx_mask" generated in Phase 3 above
- ROI Masking input moving: select "PostRx_mask" generated in Phase 3 above
- Leave all other settings at default
- click: Apply
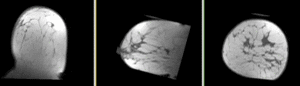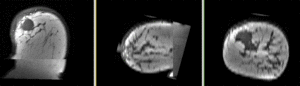
Registration Results
Discussion: Registration Challenges
- soft tissue deformations during image acquisition cause large differences in appearance
- the large tumor recession represents a significant pre/post difference in image content that will influence unmasked intensity-driven registration, which becomes a problem for the non-rigid portion of registration, particularly at higher DOF, because the registration will try to "recreate" the tumor area from the postRx image in order to match the content.
- contrast enhancement and pathology and treatment changes cause additional differences in image content
- the surface coils used cause strong differences in intensity inhomogeneity.
- we have strongly anisotropic voxel sizes with much less through-plane resolution
- resolution and FOV change between the two scans
Discussion: Key Strategies
- because of the strong changes in shape and position, we break the problem down and register each breast separately.
- we perform a bias-field correction on both images before registration
- we use the Multires version of RegisterImages for an initial affine alignment
- the nonlinear portion is then addressed with a BSpline or DiffeomorphicDemons algorithm
- because accuracy is more important than speed here, we increase the sampling rate (i.e. the number of points sampled for the BSpline registration)