Difference between revisions of "Projects:RegistrationLibrary:RegLib C10"
From NAMIC Wiki
(No difference)
| |
Revision as of 15:25, 28 May 2010
Home < Projects:RegistrationLibrary:RegLib C10Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
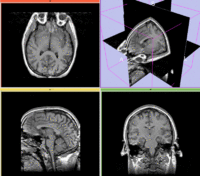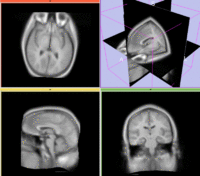
Slicer Registration Library Exampe #10: Co-registration of probabilistic tissue atlas for subsequent EM segmentation
Objective / Background
This is an example of sparse atlas co-registration. Not all atlases have an associated reference image that can be used for registration. Because the atlas represents a map of a particular tissue class probability, its contrast differs significantly from the target image.
Keywords
MRI, brain, head, inter-subject, probabilistic atlas, atlas-based segmentation
Input Data
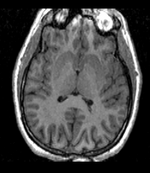
 reference/fixed : T1w axial, 1mm resolution in plane, 3mm slices
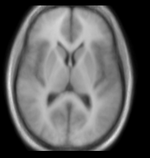
reference/fixed : T1w axial, 1mm resolution in plane, 3mm slices moving: Probabilistic Tissue atlas,
moving: Probabilistic Tissue atlas,
Methods
- build brain mask for fixed image using Skull Stripping module. Settings: 100 iterations, 20 subdivisions. New Volume: RegLib_C10_MRI_AtlasSegmentation_fixed_mask
- manually edit brain mask with Editor module. required manual fix at frontal and occipital lobe
- run Register Images , Settings:
- Fixed Image:
- Moving Image:
- Resample Image:
- Load Transform:
- Save Transform: RegLib_C10_MRI_AtlasSegmentation_Xform_Affine_wmsk
- Initialization: Centers of Mass,
- Registration: PipelineAffine
- Expected offset: 10
- Expected Rotation: 0.2
- Expected Scale: 0.1
- Expected Skew: 0.05
- Fixed Image Mask: RegLib_C10_MRI_AtlasSegmentation_fixed_mask
- Affine Max Iteration: 80
- Affine Sampling Ratio: 0.05
- (alternatively automated Affine Registration: Register Images Multires (Slicer 3.5) also produces good results
- run Deformable B-spline Registration module. Settings:
- Iterations: 20
- Grid Size: 9
- Histogram Bins: 50,
- Spatial Samples: 50000,
- initial transform: RegLib_C10_MRI_AtlasSegmentation_Xform_Affine_wmsk
- Fixed Image: RegLib_C10_MRI_AtlasSegmentation_fixed
- Fixed Image: RegLib_C10_MRI_AtlasSegmentation_moving
- Output Transform: XForm_BSpline1 -> save an output transform to then apply to other atlas data to be brought into alignment.
Registration Results
Download
- download entire package (Data,Tutorial, Solution, zip file 17 MB)
- download example & result image data (NRRD files, transforms, presets, zip file 14 MB)
- download step-by-step tutorial (PowerPoint, 3 MB)
- download registration parameter presets (zipped Slicer MRML file, 4 kB)
Link to User Guide: How to Load/Save Registration Parameter Presets
Discussion: Registration Challenges
- Because the atlas represents a probabilistic image (i.e. contains blurring from combining multiple subjects), its contrast differs significantly from the target image.
- The atlas has strong rotational misalignment that can cause difficulty for automated affine registration.
- The two images represent different anatomies, a non-rigid registration is required
Discussion: Key Strategies
- Because of the strong differences in image contrast, Mutual Information is recommended as the most robust metric.
- masking (skull stripping) is highly recommended to obtain good results for the initial affine alignment. For the 2nd stage BSpline use the full image (i.e. do NOT use the masked version) unless high-quality masks are available for both fixed & moving image. Using the crude mask created for the initial alignment for the BSpline will likely destabilize.
- because speed is not that critical, we increase the sampling rate for both affine and BSpline registration
- we also expect larger differences in scale & distortion than with regular structural scans: so we significantly (2x-3x) increase the expected values for scale and skew from the defaults.
- a good affine alignment is important before proceeding to non-rigid alignment to further correct for distortions.
Acknowledgments
- dataset provided by Killian Pohl, Ph.D.