Difference between revisions of "Projects:RegistrationLibrary:RegLib C20"
From NAMIC Wiki
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (11 intermediate revisions by one other user not shown) | |||
| Line 3: | Line 3: | ||
[[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | [[Projects:RegistrationDocumentation:UseCaseInventory|Back to Registration Use-case Inventory]] <br> | ||
| − | = <small> | + | = <small>updated for '''v4.1'''</small> [[Image:Slicer4_RegLibLogo.png|150px]] <br> Slicer Registration Library Case #20: Intra-subject whole-body PET-CT = |
=== Input === | === Input === | ||
{| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" | {| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" | ||
| Line 18: | Line 18: | ||
=== Modules === | === Modules === | ||
| − | *'''Slicer 3.6.3''' recommended modules: [ | + | *'''Slicer 3.6.3''' recommended modules: [https://www.slicer.org/wiki/Modules:BRAINSFit '''BrainsFit'''], |
===Objective / Background === | ===Objective / Background === | ||
| Line 30: | Line 30: | ||
===Download === | ===Download === | ||
| − | *Data | + | *Data: |
| − | **[[Media:RegLib_C20_Data.zip|'''RegLib C20 example data set; incl | + | **[[Media:RegLib_C20_Data.zip|'''RegLib C20 example data set; incl. solutions <small> (zip file 86 MB) </small>]] |
| − | **[[Media: | + | *ScreenCast movies |
| − | **[[ | + | **[[Media:RegLib_C20_LoadingData.mov|loading the dataset & setting up the display <small> (quicktime movie .mov 6MB) </small>]] |
| − | + | **[[Media:RegLib_C20_Registration.mov|launching registration CT-CT<small> (quicktime movie .mov 9MB) </small>]] | |
| − | + | **[[Media:RegLib_C20_Resampling.mov|resampling PET<small> (quicktime movie .mov 9MB) </small>]] | |
| − | |||
| − | |||
| − | |||
| − | [[ | ||
| − | |||
| − | |||
| − | |||
=== Procedures=== | === Procedures=== | ||
| − | *'''Phase 1''': | + | *'''Phase 1''': Load & Display |
| − | # | + | #drag & drop (or load via File menu): CT_1, CT_2, PET_1, PET_2 |
| − | ## | + | ##in the load Dialog, select ''Show Options'' and for each volume, select ''Center''. |
| − | ## | + | ##click OK |
| − | ##'' | + | #go to [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/Volumes Volumes] module |
| − | ##'' | + | ##''Active Volume'': PET_1 |
| − | ## | + | ##in the ''Display'' tab, click on the '''PET''' icon for window & level preset |
| − | ## | + | ##ditto for PET_2 |
| − | ## | + | #place CT_1 in background and PET_1 in foreground an pan between. You should see them aligned. |
| + | *'''Phase 2''': Co-register CT | ||
| + | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module] | ||
| + | ##''Fixed Image Volume'': CT_1 | ||
| + | ##''Moving Image Volume'': CT_2 | ||
| + | ##Output Settings: | ||
| + | ###''Slicer BSpline Transform": create new & rename: "Xf2_CT21" | ||
| + | ###''Slicer Linear Transform'': none | ||
| + | ###''Output Image Volume'': create new & rename: "CT_2_Xf2" | ||
| + | ##''Regstration Phases'': check boxes for ''Rigid'', "Rigid+Scale" , ''Affine'' and "BSpline" | ||
| + | ##''Main Parameters'': | ||
| + | ###''Number Of Samples'': 400,000 | ||
| + | ###''B-Spline Grid Size'': 11,11,7 | ||
| + | ##Leave all other settings at default | ||
| + | ##click: Apply; runtime ~ 5 min (MacPro QuadCore 2.4GHz) for faster performance, reduce sample points to 200,000 | ||
| + | *'''Phase 3''': Resample PET | ||
| + | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSResample BRAINSResample] module (Registration menu) | ||
| + | ##''Image To Warp'': PET_2 | ||
| + | ##''Reference Image'': PET_1 | ||
| + | ##''Output Image'': create new & rename: "PET_2_Xf2" | ||
| + | ##''Warping Parameters'' | ||
| + | ###''Warp By Transform'': Xf2_CT21'' created in Phase 2 above | ||
##leave rest at defaults | ##leave rest at defaults | ||
| − | ## | + | ##''Apply'' |
| − | # | + | #go to [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/Volumes Volumes] module |
| − | + | ##''Active Volume'': PET_2_Xf2 | |
| − | + | ##in the ''Display'' tab, click on the '''PET''' icon for window & level preset | |
| − | + | ||
| − | ##'' | + | === Registration Results=== |
| − | ##'' | + | [[Image:RegLib_C20_unregistered.gif|300px|rigid]]rigid<br> |
| − | + | [[Image:RegLib_C20_BSpline.gif|300px|BSpline registration of full volumes. 11 x 11 x 7 grid]]BSpline registration of full volumes. 11 x 11 x 7 grid<br> | |
| − | + | [[Image:RegLib_C20_BSplinePET.gif| 300px|PET overlay ; BSpline registration of full volumes. 11 x 11 x 7 grid]]PET overlay ; BSpline registration of full volumes. 11 x 11 x 7 grid [[Image:RegLib_C20_DeformGrid.png| 300px| grid overlay showing BSpline deformation ]] grid overlay showing BSpline deformation <br> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- | <!-- | ||
Latest revision as of 17:39, 10 July 2017
Home < Projects:RegistrationLibrary:RegLib C20Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
updated for v4.1 
Slicer Registration Library Case #20: Intra-subject whole-body PET-CT
Input
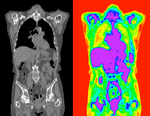
|
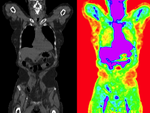
|
|
| fixed image/target | moving image |
Versions
For the Slicer 3.6 version of this case see here
Modules
- Slicer 3.6.3 recommended modules: BrainsFit,
Objective / Background
Change assessment.
Keywords
PET-CT, whole-body, change assessment
Input Data
- reference/fixed : baseline CT: 0.98 x 0.98 x 5 mm , 512 x 512 x 149; PET: 4.1 x 4.1 x 5 mm , 168 x 168 x 149
- moving: CT: 0.98 x 0.98 x 5 mm , 512 x 512 x 149; PET: 4.1 x 4.1 x 5 mm , 168 x 168 x 149
Download
- Data:
- ScreenCast movies
Procedures
- Phase 1: Load & Display
- drag & drop (or load via File menu): CT_1, CT_2, PET_1, PET_2
- in the load Dialog, select Show Options and for each volume, select Center.
- click OK
- go to Volumes module
- Active Volume: PET_1
- in the Display tab, click on the PET icon for window & level preset
- ditto for PET_2
- place CT_1 in background and PET_1 in foreground an pan between. You should see them aligned.
- Phase 2: Co-register CT
- open the General Registration (BRAINS) module
- Fixed Image Volume: CT_1
- Moving Image Volume: CT_2
- Output Settings:
- Slicer BSpline Transform": create new & rename: "Xf2_CT21"
- Slicer Linear Transform: none
- Output Image Volume: create new & rename: "CT_2_Xf2"
- Regstration Phases: check boxes for Rigid, "Rigid+Scale" , Affine and "BSpline"
- Main Parameters:
- Number Of Samples: 400,000
- B-Spline Grid Size: 11,11,7
- Leave all other settings at default
- click: Apply; runtime ~ 5 min (MacPro QuadCore 2.4GHz) for faster performance, reduce sample points to 200,000
- Phase 3: Resample PET
- open the BRAINSResample module (Registration menu)
- Image To Warp: PET_2
- Reference Image: PET_1
- Output Image: create new & rename: "PET_2_Xf2"
- Warping Parameters
- Warp By Transform: Xf2_CT21 created in Phase 2 above
- leave rest at defaults
- Apply
- go to Volumes module
- Active Volume: PET_2_Xf2
- in the Display tab, click on the PET icon for window & level preset
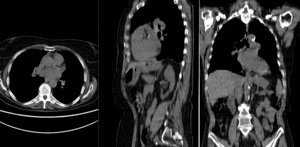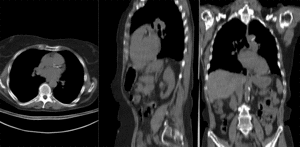
Registration Results
 rigid
rigid
 BSpline registration of full volumes. 11 x 11 x 7 grid
BSpline registration of full volumes. 11 x 11 x 7 grid
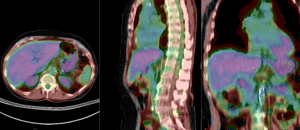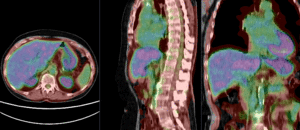 PET overlay ; BSpline registration of full volumes. 11 x 11 x 7 grid
PET overlay ; BSpline registration of full volumes. 11 x 11 x 7 grid 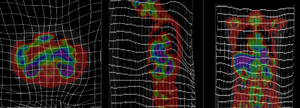 grid overlay showing BSpline deformation
grid overlay showing BSpline deformation
Discussion: Registration Challenges
- accuracy is the critical criterion here. We need the registration error (residual misalignment) to be smaller than the change we want to measure/detect. Agreement on what constitutes good alignment can therefore vary greatly.
- because of the large FOV we have strong non-rigid deformations from differences in patient/limb positions etc.
- images are large volumes (>100 MB total)
- 2 images pairs have to be aligned, i.e. the calculated transform must be applied to the second (PET) image.
Discussion: Key Strategies
- to calculate the transform, we use the images with the most accurate geometric representation and the smallest expected change, i.e. we align the follow-up CT to the baseline CT and then apply the transforms to the PET image.
- because of the non-rigid differences due to posture and breathing we will need to apply a 2-step registration with an affine alignment followed by a BSpline.