Projects:RegistrationLibrary:RegLib C46
From NAMIC Wiki
Revision as of 22:04, 28 June 2011 by Meier (talk | contribs) (Created page with 'Back to ARRA main page <br> Back to Registration main page <br> [[Projects:RegistrationDocumentation:UseCaseInv…')
Home < Projects:RegistrationLibrary:RegLib C46v3.6.1
Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
v3.6.1  Slicer Registration Library Case #46: 2D Cine MRI of Breathing Cycle
Slicer Registration Library Case #46: 2D Cine MRI of Breathing Cycle
Input
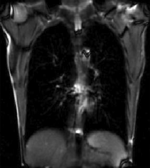
|
|
this is the time series of 2D images to be registered with the reference |
| fixed image/target | moving image |
Modules
- Slicer 3.6.1 recommended modules: BrainsFit
Objective / Background
We seek to study the motion of the lung during a breathing cycle by co-registering all of the images in a sequence with a reference, and then extract the motion from the registration displacement field.
Keywords
MRI, lung, 2D, time-series, deformation
Input Data
- reference/fixed : 2D coronal MRI 256x256
- moving: 2D coronal time series, 200 images total, each 256x256
Overall Strategy
- select one frame within the breathing cycle as reference frame
- compute non-rigid BSpline registration of all images to the reference frame
- extract the displacements of indiv. grid nodes and plot over time
Procedures
- Phase I: Pilot to determine optimal registration parameters
- load reference image and one moving image from the series
- open Registration : BrainsFit module
- Registration Phases:
- set "reference" fixed and "moving_??" as moving image
- select/check Include BSpline registration phase
- Output Settings:
- select a new transform "Slicer BSpline Transform", rename to "Xf1_moving_??"
- select a new volume "Output Image Volume, rename to "moving_??_Xf1"
- Registration Parameters: increase Number Of Samples to 200,000
- Registration Parameters: set Number Of Grid Subdivisions to 5,5,5
- Leave all other settings at default
- click: Apply; (runtime < 10 sec. on MacPro)
- adjust grid size until registration is acceptable
- Phase II: Batch Run
- open a terminal window
- via a TextEditor or prototyping/scripting software (e.g. Matlab), copy and modify the prototype line below, by changing only the moving input image:
/Applications/Slicer36/Slicer3 --launch /Applications/Slicer36/lib/Slicer3/Plugins/BRAINSFit --useBSpline --splineGridSize 5,5,5 --outputVolumePixelType short / --numberOfSamples 200000 --costMetric MMI --fixedVolume Reference/refLung_001.dcm --movingVolume Moving/Moving_001/Moving_001.dcm / --bsplineTransform Xforms/Moving_001_XfBSpl5.tfm --outputVolume MovingResampled/Moving_001_r.nrrd >> Logs/Moving_001_RegLog.txt 2>&1
- replace "/Applications/Slicer36" with your path of 3DSlicer
- create result directories MovingResampled, Logs, Xforms
- note that because input image is DICOM, and images are 2D only, each image of the time series must be in its own directory, otherwise Slicer will read them as a volume.
- paste all commands into a terminal window, or copy into a shell script and execute.
- upon completion, read the transform files with an editor and extract the displacements of interest
Registration Results
Download
- Data
- RegLib_C33_Data DWI, DTI, T1, FLAIR, Presets & solution transforms (NRRD files, zip file 174 MB)
- RegLib_C33_Data_noDWI subset of the above, contains DTI but w/o DWI and resampled DTI (NRRD files, zip file 86 MB)
- RegLib_C33_Data Transforms & Presets only contains only result transforms and parameter presets (NRRD files, zip file 30 kB)
- Presets
Discussion: Registration Challenges
- The DTI contains acquisition-related distortions (commonly EPI acquisitions) that can make automated registration difficult.
Discussion: Key Strategies
Acknowledgments
This case is the same data as case 40 from the fMRI neurosurgery data set on central.xnat.org