Difference between revisions of "Projects:ShapeAnalysisOfHippocampus"
From NAMIC Wiki
| (8 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | Back to [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|NA-MIC Collaborations]], [[Algorithm:UNC|UNC Algorithms]], [[DBP1:Dartmouth|Dartmouth DBP 1]], [[Engineering:Isomics|Isomics Engineering]] | ||
| + | __NOTOC__ | ||
= Shape Analysis of the Hippocampus = | = Shape Analysis of the Hippocampus = | ||
| − | + | Our Objective is to examine hippocampal shape in patients with schizophrenia and healthy controls. | |
| − | + | = Description = | |
| − | |||
| − | |||
| − | |||
| − | |||
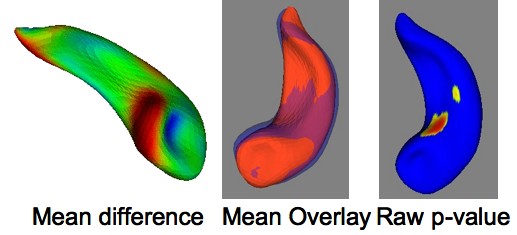
* Initial hippocampal segmentations of healthy control data were successfully processed at UNC (see average asymmetry as examplary analysis in Fig 2.) | * Initial hippocampal segmentations of healthy control data were successfully processed at UNC (see average asymmetry as examplary analysis in Fig 2.) | ||
| Line 13: | Line 11: | ||
* Have begun to apply 3D Slicer for manual segmentation. Additional subject accrual ongoing. | * Have begun to apply 3D Slicer for manual segmentation. Additional subject accrual ongoing. | ||
| − | ''' | + | '''Fig 1. Healthy Asymmetry results''' |
| + | |||
| + | [[Image:ShapeAnalysisOverviewStatsHippo05.jpg]] | ||
| − | + | = Key Investigators = | |
| − | |||
| − | |||
| − | + | * UNC Algorithms: Martin Styner, Guido Gerig | |
| − | + | * Dartmouth DBP 1: Andrew Saykin, Tara McHugh, Robert Roth, Laura Flashman | |
| − | + | * Isomics Engineering: Steve Pieper | |
| − | |||
| − | |||
| − | |||
| − | + | = Links = | |
* [[Progress_Report:Shape_Analysis|Shape Analysis Progress Report]] | * [[Progress_Report:Shape_Analysis|Shape Analysis Progress Report]] | ||
* [[DataRepository#Dartmouth_Structural.2C_Functional.2C_T2_and_DTI_Images|Data Repository for this preliminary analysis]] | * [[DataRepository#Dartmouth_Structural.2C_Functional.2C_T2_and_DTI_Images|Data Repository for this preliminary analysis]] | ||
| + | |||
| + | [[Category: Shape Analysis]] [[Category: Segmentation]] [[Category: Schizophrenia]] | ||
Latest revision as of 01:14, 28 August 2009
Home < Projects:ShapeAnalysisOfHippocampusBack to NA-MIC Collaborations, UNC Algorithms, Dartmouth DBP 1, Isomics Engineering
Shape Analysis of the Hippocampus
Our Objective is to examine hippocampal shape in patients with schizophrenia and healthy controls.
Description
- Initial hippocampal segmentations of healthy control data were successfully processed at UNC (see average asymmetry as examplary analysis in Fig 2.)
- Data has been acquired at 1.5T on 20 patients with schizophrenia and 20 healthy controls at Dartmouth.
- Have begun to apply 3D Slicer for manual segmentation. Additional subject accrual ongoing.
Fig 1. Healthy Asymmetry results
Key Investigators
- UNC Algorithms: Martin Styner, Guido Gerig
- Dartmouth DBP 1: Andrew Saykin, Tara McHugh, Robert Roth, Laura Flashman
- Isomics Engineering: Steve Pieper