Difference between revisions of "Summer2009:VCFS"
From NAMIC Wiki
| (10 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
| − | Image:PW2009-v3.png|[[2009_Summer_Project_Week| | + | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Back to Projects List]] |
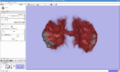
| − | Image:Anna.png|Tracts through the corpus connecting | + | Image:Anna.png|Tracts through the corpus connecting two cortical regions defined by fMRI activation. |
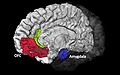
| − | Image:Image5.jpeg|ACC | + | Image:Image5.jpeg|Display of Anterior Cingulate Cortex (ACC) and Orbito-Frontal Cortex (OFC). |
| − | Image:Image11.jpeg|ACC to OFC connection | + | Image:Image11.jpeg|Population study on ACC to OFC connection using stochastic tractography showed significant FA reduction in schizophrenics. |
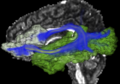
| − | Image:01040-lh-all-3D-cropped.png|IFG | + | Image:01040-lh-all-3D-cropped.png|stochastic tractography of IFG-STG (arcuate) and IFG-occipital (IOFF) connections. |
| − | Image:FA IFG-Occ scatter.jpg | + | Image:FA IFG-Occ scatter.jpg|Significant finding in left IFG-occipital connections suggests decreased integrity of fiber tracts involved in language processing in schizophrenia. |
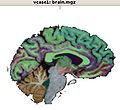
| + | Image:ScreenshotFreeSurferDeepMatterSagitalView-vcase1-2009-06-12.jpg|VCFS segmentation results to be used as seed regions for stochastic tracking. | ||
</gallery> | </gallery> | ||
| Line 16: | Line 17: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | To create an end-to-end analysis pipeline to study white matter anomalies using a stochastic tractography algorithm. | + | To create an end-to-end analysis pipeline to study white matter anomalies using a stochastic tractography algorithm in VCFS population. |
See our [[DBP2:Harvard:Brain_Segmentation_Roadmap| Roadmap]] for more details. | See our [[DBP2:Harvard:Brain_Segmentation_Roadmap| Roadmap]] for more details. | ||
| Line 32: | Line 33: | ||
* Eddy current correction | * Eddy current correction | ||
| + | We will also be testing pipeline on VCFS population, now that we have enough subjects to start a study and some encouraging gray matter segmentation results. | ||
</div> | </div> | ||
| Line 38: | Line 40: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | * Achieved ITK integration through WrapITK into the python pipeline | |
| + | ** cmake were adapted to reduce the so called 'combinatorial explosion' to a reasonable compilation cycle (<1h on my laptop) | ||
| + | ** alternative under discussion for the Slicer side: weave seems to be a better candidate | ||
| + | * Tested numpy conversion to/from VTK/ITK | ||
| + | * Ongoing : EPI distortion correction under test | ||
| + | ** Rician & EPI are cmake ready for python wrapping | ||
| + | * Even if not perfect, WrapITK allows: | ||
| + | ** to build efficient VTK/ITK pipeline - with the python benefits | ||
| + | ** to integrate easily a C++ class developed in ITK/VTK: it is just up to write a small cmake configuration file | ||
| + | ** >90% of ITK and VTK classes are usable at once | ||
| + | ** quite cumbersome to understand why people argue against it ;-) | ||
</div> | </div> | ||
Latest revision as of 14:52, 27 June 2009
Home < Summer2009:VCFSKey Investigators
- BWH: Julien de Siebenthal, Sylvain Bouix, Marek Kubicki
Objective
To create an end-to-end analysis pipeline to study white matter anomalies using a stochastic tractography algorithm in VCFS population.
See our Roadmap for more details.
Approach, Plan
Our approach involves the implementation of a stochastic tractography algorithm in python that can be used both within and outside of the slicer framework.
Our plan for the project week is to integrate diffusion weighted imaging preprocessing steps to the pipeline in particular:
- Rician Noise Removal
- EPI distortion correction
- Eddy current correction
We will also be testing pipeline on VCFS population, now that we have enough subjects to start a study and some encouraging gray matter segmentation results.
Progress
- Achieved ITK integration through WrapITK into the python pipeline
- cmake were adapted to reduce the so called 'combinatorial explosion' to a reasonable compilation cycle (<1h on my laptop)
- alternative under discussion for the Slicer side: weave seems to be a better candidate
- Tested numpy conversion to/from VTK/ITK
- Ongoing : EPI distortion correction under test
- Rician & EPI are cmake ready for python wrapping
- Even if not perfect, WrapITK allows:
- to build efficient VTK/ITK pipeline - with the python benefits
- to integrate easily a C++ class developed in ITK/VTK: it is just up to write a small cmake configuration file
- >90% of ITK and VTK classes are usable at once
- quite cumbersome to understand why people argue against it ;-)
References
- Friman, O., Farneback, G., Westin CF. A Bayesian Approach for Stochastic White Matter Tractography. IEEE Transactions on Medical Imaging, Vol 25, No. 8, Aug. 2006
- Shenton, M.E., Ngo, T., Rosenberger, G., Westin, C.F., Levitt, J.J., McCarley, R.W., Kubicki, M. Study of Thalamo-Cortical White Matter Fiber Tract Projections in Schizophrenia Using Diffusion Stochastic Tractography. Poster presented at the 46th Meeting of the American College of Neuropsychopharmacology, Boca Raton, FL, December 2007.
- Terry DP, Rausch AC, Alvarado JL, Melonakos ED, Markant D, Westin CF, Kikinis R, von Siebenthal J, Shenton ME, Kubicki M. White Matter Properties of Emotion Related Connections in Schizophrenia. Poster presented at the 2009 Mysell Poster Day, Dept. of Psychiatry, Harvard Medical School, April 2009
- Alvarado JL, Terry DP, Markant D, Ngo T, Kikinis R, Westin CF, McCarley RW, Shenton ME, Kubicki M. Study of Language-Related White Matter Tract Connections in Schizophrenia using Diffusion Stochastic Tractography. Poster presented at the 2009 Mysell Poster Day, Dept. of Psychiatry, Harvard Medical School, April 2009
- Melonakos ED, Shenton ME, Markant D, Alvarado J, Westin CF, Kubicki M. White Matter Properties of Orbitofrontal Connections in Schizophrenia. Poster being presented at the 64th Meeting of the Society of Biological Psychiatry. Vancouver, BC. May 2009.
- Kubicki, M. Khan, U., Bobrow, L., O'Donnell, L. Pieper, S. Westin, CF., Shenton, ME. New Methods for Assessing Whole Brain DTI Abnormalities in Schizophrenia. Presentation given at the International Congress of World Psychiatric Association. Florence, Italy. April 2009.
- Kubicki, M., Markant, D., Ngo, T., Westin, CF., McCarley, RW., Shenton, ME. Study of Language Related White Matter Fiber Tract Projections in Schizophrenia Using Diffusion Stochastic Tractography. Presentation given at the International Congress of World Psychiatric Association. Florence, Italy. April 2009.