Difference between revisions of "Summer project week 2010 Workflows SOA"
| (19 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2010.png|[[2010_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2010.png|[[2010_Summer_Project_Week#Projects|Projects List]] | ||
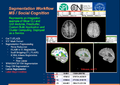
| − | Image: | + | Image:SlicerWF_PECS_SUMMARY.png|Summary of Segmentation Workflow for Social Cognition Study for MS Patients. |
| − | Image: | + | Image:SlicerWF_BPEL_Description.png|Simple Workflow Description Screenshot. BPEL Notation |
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * BWH: Alexander Zaitsev, Wendy Plesniak, Charles Guttmann, Ron Kikinis |
| − | * | + | * PECS Univeristy (Hungary): Andrea Mike |
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 21: | Line 14: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | Supplement 3D Slicer and Extensible Neuroimaging Archive Toolkit (XNAT) with workflow management system. Provide integration and interoperability between 3D Slicer, XNAT Services and 3-rd party image processing applications and high performance computing systems. Implement it as within currently developed Structural Planning and Implementation in Neurological Exploration (SPINE) System. | |
| Line 34: | Line 27: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | Our approach for analyzing diffusion tensors is summarized in the | + | Our approach for analyzing diffusion tensors is summarized in the Main Project Page reference below. |
| − | Our plan for the project week is to | + | Our plan for the project week is to present live demo for use case instances, show examples of integration Slicer3, XNAT and Cluster Computing using Service Oriented Architecture (SOA) and Workflow approach. |
| + | We also planning to quest for demanded by end-users modules that need improvement in Slicer3 (such as Brainmask Extraction), setup automation for upgraded EM Segmentation Module. We will provide on-demand tutorial for composing workflows of Slicer3 modules. | ||
</div> | </div> | ||
| Line 43: | Line 37: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | '''Prior to Project Week''': Interaction with XNAT data repository and Cluster Computing had been implemented. Several use cases had been deployed: | |
| − | + | * Emphysema Assessment: Integration of Slicer Extension, Cluster Computing, XNAT Image Repository | |
| + | * Image Preprocessing Workflow for EM Segmentation: Integration of Slicer Internal Modules, XNAT Image Repository | ||
| + | * Registration and Resampling to Common Space of Multi-Modality (PET/CT) Images for Longitudinal Studies: Integration of Slicer CLI and GUI Modules. (See [[2010_NAMIC_Project_week:LongitudinalPETSUV_Wizard | Slicer Wizard for PET/CT workflow]] for details). | ||
| + | * Complex Image Processing using Slicer3 and 3-rd Party Modules: Segmentation for Social Cognition Study for MS Patients (PECS University, Hungary [[File:SlicerWF_NAC_PECS_Workflow.pdf]] ) | ||
| + | '''During Project Week''': | ||
| + | * Adaptation of updated BRAINSFit module (Slicer 3.7 June builds) to registration pipelines | ||
| + | * Tuning Workflow for Registration and Resampling to Common Space of Multi-Modality (PET/CT) Images for Longitudinal Studies | ||
| + | * Training on Java SPECTRE skull stripping / brainmask extraction module (MIPAV version, Slicer CLI Pending release) | ||
| + | * Training on updated EM Segmenter (Slicer 3.6) | ||
| + | * Discussion of Grid Wizard (GWE) integration with workflow instances | ||
</div> | </div> | ||
| Line 53: | Line 56: | ||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
| − | This work will be delivered to the NA-MIC Kit as a | + | This work will be delivered to the NA-MIC Kit as a |
| − | #ITK Module | + | #ITK Module No |
#Slicer Module | #Slicer Module | ||
| − | ##Built-in | + | ##Built-in No |
| − | ##Extension -- commandline | + | ##Extension -- commandline Yes (Workflow Invocation Stubs) |
| − | ##Extension -- loadable | + | ##Extension -- loadable No |
| − | #Other | + | #Other -- Web Service Libraries, Workflow Composite Applications deployable on GlassFish Application Server, Rich Clients for Orchestrating Workflows and Slicer GUI |
==References== | ==References== | ||
| − | * | + | * '''Main Project Page:''' [http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerWF SlicerWF Page] |
| − | * | + | * '''Use Case Presentations:''' [http://wiki.na-mic.org/Wiki/index.php/Projects:ARRA:SlicerWF#Presentations SlicerWF Presentations] |
| − | * | + | * '''Integration with Slicer GUI:''' [[2010_NAMIC_Project_week:LongitudinalPETSUV_Wizard | Slicer Wizard for PET/CT workflow]] |
| − | |||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 15:33, 25 June 2010
Home < Summer project week 2010 Workflows SOAKey Investigators
- BWH: Alexander Zaitsev, Wendy Plesniak, Charles Guttmann, Ron Kikinis
- PECS Univeristy (Hungary): Andrea Mike
Objective
Supplement 3D Slicer and Extensible Neuroimaging Archive Toolkit (XNAT) with workflow management system. Provide integration and interoperability between 3D Slicer, XNAT Services and 3-rd party image processing applications and high performance computing systems. Implement it as within currently developed Structural Planning and Implementation in Neurological Exploration (SPINE) System.
Approach, Plan
Our approach for analyzing diffusion tensors is summarized in the Main Project Page reference below.
Our plan for the project week is to present live demo for use case instances, show examples of integration Slicer3, XNAT and Cluster Computing using Service Oriented Architecture (SOA) and Workflow approach. We also planning to quest for demanded by end-users modules that need improvement in Slicer3 (such as Brainmask Extraction), setup automation for upgraded EM Segmentation Module. We will provide on-demand tutorial for composing workflows of Slicer3 modules.
Progress
Prior to Project Week: Interaction with XNAT data repository and Cluster Computing had been implemented. Several use cases had been deployed:
- Emphysema Assessment: Integration of Slicer Extension, Cluster Computing, XNAT Image Repository
- Image Preprocessing Workflow for EM Segmentation: Integration of Slicer Internal Modules, XNAT Image Repository
- Registration and Resampling to Common Space of Multi-Modality (PET/CT) Images for Longitudinal Studies: Integration of Slicer CLI and GUI Modules. (See Slicer Wizard for PET/CT workflow for details).
- Complex Image Processing using Slicer3 and 3-rd Party Modules: Segmentation for Social Cognition Study for MS Patients (PECS University, Hungary File:SlicerWF NAC PECS Workflow.pdf )
During Project Week:
- Adaptation of updated BRAINSFit module (Slicer 3.7 June builds) to registration pipelines
- Tuning Workflow for Registration and Resampling to Common Space of Multi-Modality (PET/CT) Images for Longitudinal Studies
- Training on Java SPECTRE skull stripping / brainmask extraction module (MIPAV version, Slicer CLI Pending release)
- Training on updated EM Segmenter (Slicer 3.6)
- Discussion of Grid Wizard (GWE) integration with workflow instances
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a
- ITK Module No
- Slicer Module
- Built-in No
- Extension -- commandline Yes (Workflow Invocation Stubs)
- Extension -- loadable No
- Other -- Web Service Libraries, Workflow Composite Applications deployable on GlassFish Application Server, Rich Clients for Orchestrating Workflows and Slicer GUI
References
- Main Project Page: SlicerWF Page
- Use Case Presentations: SlicerWF Presentations
- Integration with Slicer GUI: Slicer Wizard for PET/CT workflow