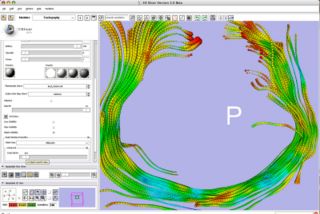
Projects/Diffusion/2007 Project Week Slicer3 Tractography Editor
 Return to Project Week Main Page |
Key Investigators
- BWH: Lauren O'Donnell, Raul San Jose, Alex Yarmarkovich
Objective
Initial development of tractography editing tools. These will eventually include region of interest based selection of fiber bundles in slicer 3, conversion of fiber bundles to voxels, etc.
Approach, Plan
Build on existing MRML tractography display infrastructure to allow ROI selection of fibers.
Software for the MRML infrastructure and tractography display has been implemented, and is undergoing further improvement. Software for whole brain seeding is being developed in slicer3 as a concurrent programming week project.
Our plan for the project week is to create a CLP module to do region of interest selection of whole brain tractography.
Progress
A CLP module, Tractography->Editor->ROISelect, has been created.
Also, another CLP module for probing volumes with models ("painting" scalars on models) was created and checked into Slicer3 svn.
There were discussions about simpler API for display of data like tractography (that needs pipeline(s) to convert to displayable polydata) with Alex, Steve, Raul, and Demian.
We also planned to enable future display of tractography with non-tensor or multiple tensor data attached (with Raul and Demian).
We fixed a bug with MRML scene reading for fiber bundles, so that is working correctly now.