Projects/Slicer3/2007 Project Week Support for electron microscopy
 Return to Project Week Main Page |
Key Investigators
- UCSD/NCMIR: W. Bryan Smith, Mark Ellisman
- Isomics: Steve Pieper
Objective
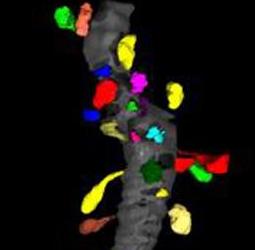
Dendritic spines are the primary sites of excitatory neuronal communication in the brain. It is of fundamental interest to cellular neurobiologists to understand how the number, size, and shape of dendritic spines varies across brain regions, and as a function of experimental treatment within a given region. We would like to assess morphological properties of spines using Slicer.
Approach, Plan
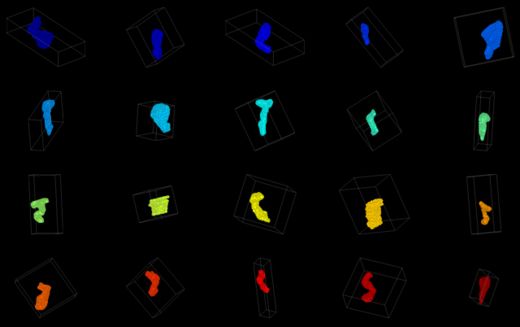
Spines have been manually segmented from tilt-series electron tomograms. The spines need to be registered to a common coordinate system, and then a variety of measurements must be made on the size and shape of each spine in an effort to cluster the spines according to their morphologies.
The plan for the project week is to first try out 3D skeletonization filters in ITK to define the medial axis of each spine.
Progress
- Attempted itk::BinaryThinningImageFilter (Doxygen Link), but it creates planes rather than lines in parts of some spines.
- Obtained code from Martin Styner (via Stephen Aylward). Will implement and see if works.
- Suggestion to try CorCTA module in Slicer2; Crashes
References
- Insert some refs here