Difference between revisions of "ProjectWeek200706:ContrastingTractographyMeasures"
| (61 intermediate revisions by 9 users not shown) | |||
| Line 6: | Line 6: | ||
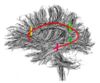
|[[Image:InternalCapsule.jpg|thumb|left|100px|Internal Capsule]] | |[[Image:InternalCapsule.jpg|thumb|left|100px|Internal Capsule]] | ||
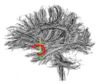
|[[Image:Uncinate.jpg|thumb|left|100px|Uncinate Fasciculus]] | |[[Image:Uncinate.jpg|thumb|left|100px|Uncinate Fasciculus]] | ||
| + | <br> | ||
| + | <br> | ||
|} | |} | ||
| + | ---- | ||
| + | |||
| + | =='''THIS PAGE IS BEING USED FOR PROJECT MANAGEMENT- LINK ALL IMPORTANT WIKI PAGES IN THE PROGRESS SECTION''' == | ||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
__NOTOC__ | __NOTOC__ | ||
| − | |||
===Key Investigators=== | ===Key Investigators=== | ||
| − | * BWH: Marek Kubicki, Doug Markant, Doug Terry | + | * BWH: Marek Kubicki (Core 3), Carl-Fredrik Westin (NAC collaborator), Lauren O'Donnell (Core 1), Sonia Pujol (Core 5), Doug Markant (Core 3), Doug Terry (Core 3), Katharina Quintus (Core 3), Jorge Alvarado (Core 3), Tri Ngo (Core 3), Sylvain Bouix (Core 3), Marc Niethammer (Core 3), |
| − | * MGH: Bruce Fischl, Denis Jen | + | * MGH: Randy Gollub (Core 5), Bruce Fischl (Core 1), Denis Jen (Core 1), Anastasia Yendiki, Karl Helmer (Morphometry BIRN collaborator) |
| − | * Utah: Tom Fletcher, Ross Whitaker, Guido Gerig, | + | * Utah: Tom Fletcher (Core 1), Ross Whitaker (Cores 1 & 5), Guido Gerig (Cores 1 & 5), Casey Goodlett (Core 1), Sylvain Gouttard |
| − | * UNC: | + | * UNC: Martin Styner (Core 1) |
| − | * GA Tech: | + | * GA Tech: John Melonakos (Core 1), Vandana Mohan (Core 1), Allen Tannenbaum (Core 1) |
| − | * MIT: | + | * MIT: Polina Golland (Core 1) |
| + | * UIowa: Vince Magnotta (Collaborator) | ||
| + | * UNM: Jeremy Bockholt (Core 3/Collaborator) | ||
| + | * UCLA: Nathan Hageman (Core 1) | ||
| + | * UCI: Jim Fallon (Core 3, alumnus), Adrian Preada (Core 3, alumnus) | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 25: | Line 37: | ||
* There are many outstanding questions in this domain that we agree are interesting and worth considering such as: | * There are many outstanding questions in this domain that we agree are interesting and worth considering such as: | ||
| − | 1. What benchmarks should be used to assess performance of a | + | 1. What benchmarks should be used to assess performance of a NA-MIC Toolkit algorithm? |
<p>2. How can we assess the performance of an algorithm if we have no access to the ground truth of what it is measuring (e.g. the white matter of the brain with tractography)?</P> | <p>2. How can we assess the performance of an algorithm if we have no access to the ground truth of what it is measuring (e.g. the white matter of the brain with tractography)?</P> | ||
<p>3. What statistical methods are most appropriate for quanitfying and testing significance of these assessments? </p> | <p>3. What statistical methods are most appropriate for quanitfying and testing significance of these assessments? </p> | ||
| − | The answers to these questions will vary depending on the specific algorithm and its application. The group agreed that the best way to proceed was to chose one very specific example that is highly relevant to the | + | The answers to these questions will vary depending on the specific algorithm and its application. The group agreed that the best way to proceed was to chose one very specific example that is highly relevant to the NA-MIC work to date and focus efforts on that. The methods that arise from this can then be applied to additional areas. |
We agreed to begin by studying the results obtained by applying each of the tractography tools to a single dataset and then gathering to present the results to one another and discuss how best to quantify the similarities and differences. | We agreed to begin by studying the results obtained by applying each of the tractography tools to a single dataset and then gathering to present the results to one another and discuss how best to quantify the similarities and differences. | ||
| Line 37: | Line 49: | ||
1. To write up the results for publication with all of us as contributing authors. | 1. To write up the results for publication with all of us as contributing authors. | ||
<p>2. To make the dataset and our analyzed results available to the scientific community </p> | <p>2. To make the dataset and our analyzed results available to the scientific community </p> | ||
| − | <p>3. To use our findings to begin to establish benchmarking methods for the | + | <p>3. To use our findings to begin to establish benchmarking methods for the NA-MIC toolkit.</p> |
</div> | </div> | ||
| Line 46: | Line 58: | ||
Details: | Details: | ||
| − | * | + | * The first data set was provided by Marek Kubicki and used to generate the results for the Santa Fe Workshop. Cohort includes 5 subjects with schizophrenia and 5 matched healthy controls that are de-identified and not marked as to diagnosis. Each subject has a 3T high resolution DTI scan, structural MRI scan and expert generated Regions of Interest (ROIs) for each subject that are needed for tract definition. Results from Santa Fe drove the decision to switch to a data set that had two study visits per subject to enable reliability estimates. |
| + | |||
| + | * The second dataset was provided by Randy Gollub and the MIND Clinical Imaging Consortium (MCIC). Cohort includes 10 healthy control subjects scanned twice. Each subject has a 1.5 T DTI scan, structural MRI scans and fMRI scans. | ||
| + | |||
| + | * '''Acquisition parameters''' for both data sets can be found here: [[Projects/Diffusion/2007_Project_Week_Contrasting Tractography Measures/Acquisition Parameters|Acquisition Parameters]]. | ||
| + | |||
| + | * The '''12 tracts''' being studied are the cingulum bundle, the uncinate fasiculus, the fornix, the internal capsule, and the arcuate fasiculus on the left and right sides, as well as the corpus callosum ( forceps major and forceps minor). To see how the first set of ROI's were defined, click here: [[Projects/Diffusion/2007_Project_Week_Contrasting Tractography Measures/ROI_Definitions|ROI Definitions]]. | ||
| + | |||
| + | * The '''file naming convention''' for uploading the results on the birn portal is as follows: | ||
| + | |||
| + | ** MIND subjects data: '''tractName_toolName_caseNumber_visitNumber_tracts.vtk''': ''leftCingulum_FiberViewer_M87101083_visit1_tracts.vtk rightCingulum_FiberViewer_M87101083_visit1_tracts.vtk''. | ||
| + | Please group the left and right side together in a tgz archive: ''cingulum_FiberViewer_M87101083_visit1_tracts.tgz'' | ||
| − | * | + | ** Phantom data: '''tractName_toolName_phantomName_tracts.vtk''' |
| + | ''tract1inf_slicerROI_teemPhantom_tracts.vtk'' | ||
| + | ''tract2sup_slicerROI_teemPhantom_tracts.vtk'' | ||
| − | * Each tool developer is responsible for downloading and analyzing the data, optimizing their own algorithm as needed. Keep careful notes on your final processing methods as you will need to teach them to Sonia Pujol | + | * Each tool developer is responsible for downloading and analyzing the data, optimizing their own algorithm as needed. Keep careful notes on your final processing methods as you will need to teach them to Sonia Pujol at the Santa Fe retreat. She will repeat the analysis independently using all the tools herself on the data, these are the result that will be included in summary manuscript(s). |
| − | * Metrics to be collected | + | * Metrics to be collected will be finalized by this group in an upcoming T-con. Suggestions include measure of FA along the tract, size and/or volume of tract, spatial localization of tract, measure of connectivity. Perhaps also some way to look at group results towards the goal of being able to make statements about differences in health and disease? |
* Participating algorithms (and tester) include: | * Participating algorithms (and tester) include: | ||
| − | # | + | # Fiber tracking the UNC way (Guido Gerig, Casey Goodlett or his designee) |
| − | # Slicer tract tool ( | + | # Slicer tract tool (Sonia Pujol/Doug Terry/Marek Kubicki) |
| − | |||
# Volumetric connectivity (Ross Whitaker/Tom Fletcher) | # Volumetric connectivity (Ross Whitaker/Tom Fletcher) | ||
| − | # | + | # [[Projects/Diffusion/Contrasting_Tractography_Measures#GATech|Geodesic Tractography Segmentation]] (Allen Tannenbaum/John Melonakos) |
| − | |||
# GTRACT (Vince Magnotta) | # GTRACT (Vince Magnotta) | ||
| − | # | + | # Stochastic Tractography (Tri Ngo/Carl-Fredrik Westin) |
| + | # Fluid Mechanics Tractography (Nathan Hageman/Arthur Toga) | ||
| + | |||
| + | In preparation for the upcoming T-con, each algorithm team needs to update this wiki page that describes all the participating [[Projects/Diffusion/Contrasting_Tractography_Measures | tractography algorithms ]] with proposed input and outputs for comparison. UNC and Iowa are done already. | ||
| − | * We will then hold a 2 day | + | * We will then hold a 2 day conference in [[SanteFe.Tractography.Conference| Santa Fe, NM on Oct 1-2]]. This time window avoids MICCAI, BIRN, SFN, New Mexico's ballonfest, and the Jewish High Holidays. The workshop/retreat will include a presentation from each group of their results AND a recommendation for how to statstically compare and quantify similarities and differences within and across tools. The outcome of the workshop/retreat will be a set of final agreed upon measurement metrics and method for compare and quantify similarities and differences within and across tools. And the training of Sonia Pujol to run each of the analysis tools. |
</div> | </div> | ||
| Line 81: | Line 107: | ||
:dwi-EdCor (raw diffusion data eddy current corrected) | :dwi-EdCor (raw diffusion data eddy current corrected) | ||
:dougt_DTI_ROI (directory of labelmaps) | :dougt_DTI_ROI (directory of labelmaps) | ||
| + | |||
| + | |||
| + | Interest in project confirmed during Project Week and dates for [[SanteFe.Tractography.Conference| Sante Fe meeting]] agreed upon by group. Next step is to convene a planning T-con to organize the final meeting preparations including how to share results in advance, who will attend and clarify roles for participants. <br> | ||
| + | Meeting Notes are here [[July31T-con | SanteFe Tractography Conference July 31st Planning T-con notes Page]] | ||
| + | |||
| + | Summary of Santa Fe Workshop and action plans, including data set description and instructions for downloading data are here [[TractographyWkshop_Core1_ActionPlan]] | ||
| + | |||
| + | [[Tractography_Study_Telephone_Conference_Nov_16_2007| Notes from Tractography Study TCon November 16, 2007 - 2 pm EST]] | ||
| + | |||
| + | AHM 2008 DTI VALIDATION WORKSHOP [[2008_Winter_Project_Week_Tractography| AGENDA]] AND [[2008_Winter_Project_Week_Tractography_Meeting_Notes | MEETING NOTES]] | ||
| + | |||
| + | [[DTI_Validation_Telephone_Conferences_2008 | Notes from the DTI Validation TCons ]] | ||
| + | |||
| + | [http://www.na-mic.org/Wiki/index.php/AHM_2009:DTI_Breakout AHM 2009 DTI VALIDATION SESSION ] | ||
| + | |||
| + | ''' Presentation of the preliminary results of the NA-MIC DTI Validation Study ''' at the [http://www.ismrm.org/09/ Annual Meeting of the International Society for Magnetic Resonance in Imaging (ISMRM 2009)] | ||
| + | |||
| + | 'Preliminary Results on the use of STAPLE for evaluating DT-MRI tractography in the absence of ground truth.' S. Pujol, C-F. Westin, R. Whitaker, G. Gerig, T. Fletcher, V. Magnotta, S. Bouix, R. Kikinis, W. M. Wells III, and R. Gollub. In Proceedings ISMRM 2009, Apr 18-24, 2009. Honolulu, Hawaii. | ||
| + | |||
</div> | </div> | ||
| Line 87: | Line 132: | ||
</div> | </div> | ||
| + | |||
| + | |||
==Additional Information== | ==Additional Information== | ||
*[[DBP2:Harvard|Harvard DBP (II) Homepage]] | *[[DBP2:Harvard|Harvard DBP (II) Homepage]] | ||
| + | |||
| + | == This Project's Historical Background== | ||
| + | *[[Training:Tractography.Validation | Tractography Validation]] | ||
| + | |||
| + | =Sponsors= | ||
| + | <gallery> | ||
| + | Image:NAMIC 380x463.jpg|[http://www.na-mic.org National Alliance for Medical Image Computing] | ||
| + | Image:Nac.png|[http://nac.spl.harvard.edu/nac Neuroimage Analysis Center] | ||
| + | Image:NCIGTlogo.gif|[http://www.ncigt.org National Center for Image Guided therapy] | ||
| + | Image:LogoBIRN.jpg|[http://www.nbirn.net/ BIRN] | ||
| + | </gallery> | ||
Latest revision as of 19:00, 15 May 2009
Home < ProjectWeek200706:ContrastingTractographyMeasures Return to Project Week Main Page |
|
THIS PAGE IS BEING USED FOR PROJECT MANAGEMENT- LINK ALL IMPORTANT WIKI PAGES IN THE PROGRESS SECTION
Key Investigators
- BWH: Marek Kubicki (Core 3), Carl-Fredrik Westin (NAC collaborator), Lauren O'Donnell (Core 1), Sonia Pujol (Core 5), Doug Markant (Core 3), Doug Terry (Core 3), Katharina Quintus (Core 3), Jorge Alvarado (Core 3), Tri Ngo (Core 3), Sylvain Bouix (Core 3), Marc Niethammer (Core 3),
- MGH: Randy Gollub (Core 5), Bruce Fischl (Core 1), Denis Jen (Core 1), Anastasia Yendiki, Karl Helmer (Morphometry BIRN collaborator)
- Utah: Tom Fletcher (Core 1), Ross Whitaker (Cores 1 & 5), Guido Gerig (Cores 1 & 5), Casey Goodlett (Core 1), Sylvain Gouttard
- UNC: Martin Styner (Core 1)
- GA Tech: John Melonakos (Core 1), Vandana Mohan (Core 1), Allen Tannenbaum (Core 1)
- MIT: Polina Golland (Core 1)
- UIowa: Vince Magnotta (Collaborator)
- UNM: Jeremy Bockholt (Core 3/Collaborator)
- UCLA: Nathan Hageman (Core 1)
- UCI: Jim Fallon (Core 3, alumnus), Adrian Preada (Core 3, alumnus)
Objective
A new initiative has begun in response to a shared vision among Cores 1, 3 and 5 that the field of medical image analysis would be well served by work in the area of validation, calibration and assessment of reliability. Discussions have continued among our participants since then and as a result a plan for the initial work on this front has been articulated.
- There are many outstanding questions in this domain that we agree are interesting and worth considering such as:
1. What benchmarks should be used to assess performance of a NA-MIC Toolkit algorithm?
2. How can we assess the performance of an algorithm if we have no access to the ground truth of what it is measuring (e.g. the white matter of the brain with tractography)?
3. What statistical methods are most appropriate for quanitfying and testing significance of these assessments?
The answers to these questions will vary depending on the specific algorithm and its application. The group agreed that the best way to proceed was to chose one very specific example that is highly relevant to the NA-MIC work to date and focus efforts on that. The methods that arise from this can then be applied to additional areas.
We agreed to begin by studying the results obtained by applying each of the tractography tools to a single dataset and then gathering to present the results to one another and discuss how best to quantify the similarities and differences.
- Final goals are:
1. To write up the results for publication with all of us as contributing authors.
2. To make the dataset and our analyzed results available to the scientific community
3. To use our findings to begin to establish benchmarking methods for the NA-MIC toolkit.
Approach
Details:
- The first data set was provided by Marek Kubicki and used to generate the results for the Santa Fe Workshop. Cohort includes 5 subjects with schizophrenia and 5 matched healthy controls that are de-identified and not marked as to diagnosis. Each subject has a 3T high resolution DTI scan, structural MRI scan and expert generated Regions of Interest (ROIs) for each subject that are needed for tract definition. Results from Santa Fe drove the decision to switch to a data set that had two study visits per subject to enable reliability estimates.
- The second dataset was provided by Randy Gollub and the MIND Clinical Imaging Consortium (MCIC). Cohort includes 10 healthy control subjects scanned twice. Each subject has a 1.5 T DTI scan, structural MRI scans and fMRI scans.
- Acquisition parameters for both data sets can be found here: Acquisition Parameters.
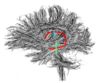
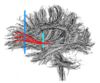
- The 12 tracts being studied are the cingulum bundle, the uncinate fasiculus, the fornix, the internal capsule, and the arcuate fasiculus on the left and right sides, as well as the corpus callosum ( forceps major and forceps minor). To see how the first set of ROI's were defined, click here: ROI Definitions.
- The file naming convention for uploading the results on the birn portal is as follows:
- MIND subjects data: tractName_toolName_caseNumber_visitNumber_tracts.vtk: leftCingulum_FiberViewer_M87101083_visit1_tracts.vtk rightCingulum_FiberViewer_M87101083_visit1_tracts.vtk.
Please group the left and right side together in a tgz archive: cingulum_FiberViewer_M87101083_visit1_tracts.tgz
- Phantom data: tractName_toolName_phantomName_tracts.vtk
tract1inf_slicerROI_teemPhantom_tracts.vtk tract2sup_slicerROI_teemPhantom_tracts.vtk
- Each tool developer is responsible for downloading and analyzing the data, optimizing their own algorithm as needed. Keep careful notes on your final processing methods as you will need to teach them to Sonia Pujol at the Santa Fe retreat. She will repeat the analysis independently using all the tools herself on the data, these are the result that will be included in summary manuscript(s).
- Metrics to be collected will be finalized by this group in an upcoming T-con. Suggestions include measure of FA along the tract, size and/or volume of tract, spatial localization of tract, measure of connectivity. Perhaps also some way to look at group results towards the goal of being able to make statements about differences in health and disease?
- Participating algorithms (and tester) include:
- Fiber tracking the UNC way (Guido Gerig, Casey Goodlett or his designee)
- Slicer tract tool (Sonia Pujol/Doug Terry/Marek Kubicki)
- Volumetric connectivity (Ross Whitaker/Tom Fletcher)
- Geodesic Tractography Segmentation (Allen Tannenbaum/John Melonakos)
- GTRACT (Vince Magnotta)
- Stochastic Tractography (Tri Ngo/Carl-Fredrik Westin)
- Fluid Mechanics Tractography (Nathan Hageman/Arthur Toga)
In preparation for the upcoming T-con, each algorithm team needs to update this wiki page that describes all the participating tractography algorithms with proposed input and outputs for comparison. UNC and Iowa are done already.
- We will then hold a 2 day conference in Santa Fe, NM on Oct 1-2. This time window avoids MICCAI, BIRN, SFN, New Mexico's ballonfest, and the Jewish High Holidays. The workshop/retreat will include a presentation from each group of their results AND a recommendation for how to statstically compare and quantify similarities and differences within and across tools. The outcome of the workshop/retreat will be a set of final agreed upon measurement metrics and method for compare and quantify similarities and differences within and across tools. And the training of Sonia Pujol to run each of the analysis tools.
Progress
Data can be found on BIRN in the following directory:
/home/Projects/NAMIC__0003/Files/PNL/3T_data_sample
with subdirectories:
- dwi (raw diffusion data)
- dwi-EdCor (raw diffusion data eddy current corrected)
- dougt_DTI_ROI (directory of labelmaps)
Interest in project confirmed during Project Week and dates for Sante Fe meeting agreed upon by group. Next step is to convene a planning T-con to organize the final meeting preparations including how to share results in advance, who will attend and clarify roles for participants.
Meeting Notes are here SanteFe Tractography Conference July 31st Planning T-con notes Page
Summary of Santa Fe Workshop and action plans, including data set description and instructions for downloading data are here TractographyWkshop_Core1_ActionPlan
Notes from Tractography Study TCon November 16, 2007 - 2 pm EST
AHM 2008 DTI VALIDATION WORKSHOP AGENDA AND MEETING NOTES
Notes from the DTI Validation TCons
AHM 2009 DTI VALIDATION SESSION
Presentation of the preliminary results of the NA-MIC DTI Validation Study at the Annual Meeting of the International Society for Magnetic Resonance in Imaging (ISMRM 2009)
'Preliminary Results on the use of STAPLE for evaluating DT-MRI tractography in the absence of ground truth.' S. Pujol, C-F. Westin, R. Whitaker, G. Gerig, T. Fletcher, V. Magnotta, S. Bouix, R. Kikinis, W. M. Wells III, and R. Gollub. In Proceedings ISMRM 2009, Apr 18-24, 2009. Honolulu, Hawaii.