Difference between revisions of "NA-MIC External Collaborations"
| Line 50: | Line 50: | ||
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:Virtual Colonoscopy Auto Detection - Yoshida.png|thumb|left|300px]] |
| | | | | | ||
Revision as of 15:29, 10 July 2009
Home < NA-MIC External CollaborationsBack to NA-MIC Collaborations
Contents
- 1 Projects funded by "Collaborations with NCBC PAR"
- 1.1 PAR-05-063: R01EB005973 Automated FE Mesh Development
- 1.2 PAR-07-249: R01AA016748 Measuring Alcohol and Stress Interaction with Structural and Perfusion MRI
- 1.3 PAR-05-063: R01CA124377 An Integrated System for Image-Guided Radiofrequency Ablation of Liver Tumors
- 1.4 PAR-07-249: R01EB006733 Development and Dissemination of Robust Brain MRI Measurement Tools
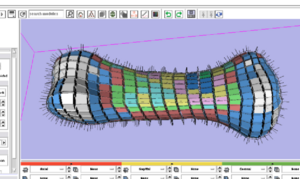
- 1.5 PAR-07-249: R01CA131718 NA-MIC virtual colonoscopy
- 1.6 PAR-07-249: 1R01 EB008171-01A1 3D Shape Analysis for Computational Anatomy
- 2 Additional External Collaborations
- 2.1 PAR-05-057: BRAINS Morphology and Image Analysis
- 2.2 Vascular Modeling Toolkit Collaboration
- 2.3 Children's Pediatric Cardiology Collaboration with SCI/SPL/Northeastern
- 2.4 NA-MIC Collaboration with NITRC
- 2.5 NA-MIC Collaboration with NAC
- 2.6 NA-MIC Collaboration with NCIGT
- 2.7 NA-MIC Collaboration with Research and Development Project on Intelligent Surgical Instruments
- 2.8 Real Time Computer Simulation of Human Soft Organ Deformation for Computer Assisted Surgery
- 2.9 Real-Time Computing for Image Guided Neurosurgery
- 2.10 NA-MIC support for Harvard CTSC Translational Imaging Consortium
- 2.11 Stanford Simbios
- 3 Broader Community Involvement of NA-MIC Investigators
Projects funded by "Collaborations with NCBC PAR"
This section describes external collaborations with NA-MIC that are funded by NIH under the "Collaboration with NCBC" PAR. (Details for this funding mechanism are provided here).
|
PAR-05-063: R01EB005973 Automated FE Mesh DevelopmentThis project is funded under an NCBC collaboration grant to PIs Nicole Grosland and Vincent Magnotta at UIowa. The goal of this project is to integrate and expand methods to automate the development of specimen- / patient-specific finite element (FE) models into the NA-MIC kit. More... | |
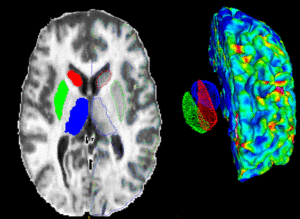
PAR-07-249: R01AA016748 Measuring Alcohol and Stress Interaction with Structural and Perfusion MRIThis project is funded under an NCBC collaboration grant to PIs James Daunais, Robert Kraft, and Chris Wyatt. The goal of this project is to examine the the effects of chronic alcohol self- administration on brain structure and function the monkey brain. MRI image analysis tools from the NA-MIC kit will be adapted for use with the monkey brain datasets. More... | |
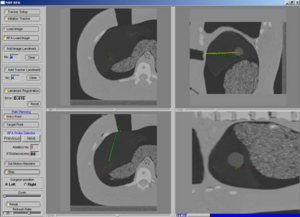
PAR-05-063: R01CA124377 An Integrated System for Image-Guided Radiofrequency Ablation of Liver TumorsThis project is funded under an NCBC collaboration grant to PI Kevin Cleary at Georgetown University. The goal of this project is to develop and validate an integrated system based on open source software for improved visualization and probe placement during radiofrequency ablation (RFA) of liver tumors.More... | |
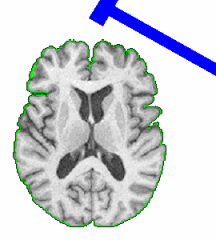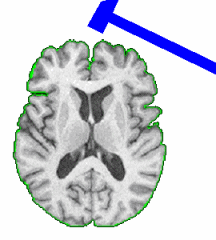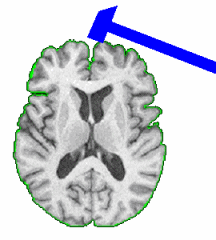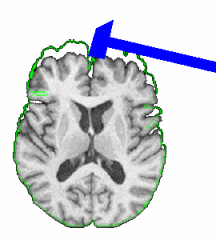
PAR-07-249: R01EB006733 Development and Dissemination of Robust Brain MRI Measurement ToolsThis project is funded under an NCBC collaboration grant to PI Dinggang Shen at UNC-Chapel Hill. The goal of this project is to develop and widely distribute a software package for robust measurement of brain structures in MR images using computational neuroanatomy methods.More... | |
| [[Image:|thumb|left|300px]] |
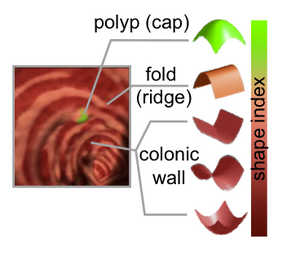
PAR-07-249: R01CA131718 NA-MIC virtual colonoscopyThis project is funded under an NCBC collaboration grant to PI Hiroyuki Yoshida. The goal of this project is to More... |
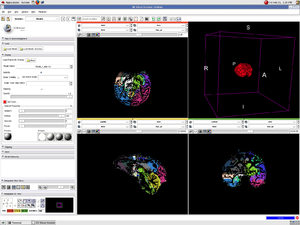
PAR-07-249: 1R01 EB008171-01A1 3D Shape Analysis for Computational AnatomyThis project is funded under an NCBC collaboration grant to PI Michael Miller JHU (with Joe Hennessey) |
Additional External Collaborations
This section describes external collaborations with NA-MIC that are funded by other mechanisms:
PAR-05-057: BRAINS Morphology and Image AnalysisThis project is a funded under a Continued Development and Maintenance of Software grant to PIs Vincent Magnotta, Hans Johnson, Jeremy Bockholt, and Nancy Andreasen at the University of Iowa. The goal of this project is to update the BRAINS image analysis software developed at the University of Iowa. More... | |
Vascular Modeling Toolkit CollaborationSlicer as a platform for segmentation and geometric analysis of vascular segments and image-based computational fluid dynamics (CFD). More... Collaboration with Luca Antiga of the Mario Negri Institute. | |
Children's Pediatric Cardiology Collaboration with SCI/SPL/NortheasternCollaboration with John Triedman, Matt Jolley, Dana Brooks, SCI. | |
NA-MIC Collaboration with NITRCThe NA-MIC Project is working to make NA-MIC neuroimaging software available through the NITRC web site. Supplemental support is helping to create the Slicer3 Loadable Modules project so that slicer plugins can be hosted on NITRC, allowing greater scalability for developers and users of Slicer. | |
NA-MIC Collaboration with NACNAC, the neuroimage analysis center, is a national resource center. NAC is relying on the NA-MIC kit for its general software environment. The mission of NAC is to develop novel concepts for the analysis of images of the brain and develop and disseminate tools based on those concepts. | |
NA-MIC Collaboration with NCIGTNCIGT is leveraging the NA-MIC kit as a platform for developing dedicated IGT capabilities.
| |
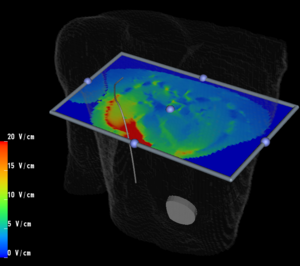
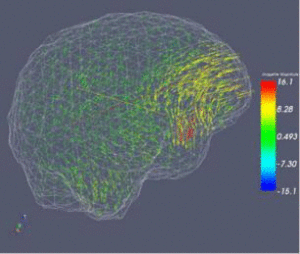
NA-MIC Collaboration with Research and Development Project on Intelligent Surgical InstrumentsIntelligent Surgical Instruments Projects uses Open-source software engineering tools developed by NA-MIC, and leverage it to surgical robotics.
| |
Real Time Computer Simulation of Human Soft Organ Deformation for Computer Assisted SurgeryReal Time Computer Simulation of Human Soft Organ Deformation for Computer Assisted Surgery. | |
Real-Time Computing for Image Guided NeurosurgeryUsing the Tera Grid to implement mesh-based non-rigid registration for Neurosurgery. | |

|

NA-MIC support for Harvard CTSC Translational Imaging ConsortiumThe Harvard CTSC Translational Imaging Consortium is using NA-MIC communication tools to facilitate the rapid deployment of expertise in medical imaging acquisition, analysis and visualization to clinical translational investigators.
|
|
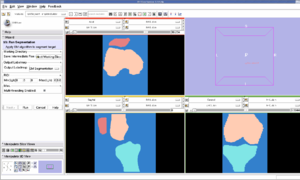
Stanford SimbiosOur sister NCBC at Stanford, dedicated to biomedical simulation, is working to adapt NA-MIC image analysis routines generate simulation models directly from MRI scans.
|
Broader Community Involvement of NA-MIC Investigators
This section describes external efforts that are not specifically NA-MIC collaborations but share personnel and/or technology:
File:Placeholder.gif 300px |
PlaceholderPlaceholder |
File:Placeholder.gif 300px |
PlaceholderPlaceholder
|