Difference between revisions of "2008 Summer Project Week:ProstateSegReg"
From NAMIC Wiki
(New page: {| |thumb|320px|Return to [[2008_Summer_Project_Week|Project Week Main Page ]] |} __NOTOC__ ===Key Investigators=== * Allen Tannenbaum, Yi Gao, Georgia Tec...) |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:ProjectWeek-2008.png|thumb|320px|Return to [[2008_Summer_Project_Week|Project Week Main Page]] ]] | |[[Image:ProjectWeek-2008.png|thumb|320px|Return to [[2008_Summer_Project_Week|Project Week Main Page]] ]] | ||
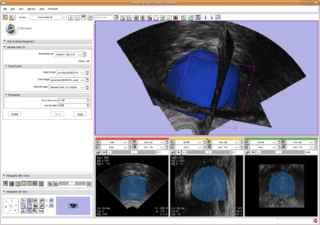
| + | |[[Image:SlicerRandomWalkProstate.png |thumb|320px| Segmentation for prostate. Random Walk algorithm as a slicer command line module. ]] | ||
|} | |} | ||
| Line 8: | Line 9: | ||
* Allen Tannenbaum, Yi Gao, Georgia Tech | * Allen Tannenbaum, Yi Gao, Georgia Tech | ||
* Gabor Fichtinger, Purang Abolmaesumi, Siddharth Vikal, David Gobbi, Queen's University | * Gabor Fichtinger, Purang Abolmaesumi, Siddharth Vikal, David Gobbi, Queen's University | ||
| + | * Luis Ibanez, Kitware | ||
| Line 15: | Line 17: | ||
<h1>Objective</h1> | <h1>Objective</h1> | ||
| − | + | * To integrate current segmentation algorithm into the software environment being used by Core 3 people. | |
| − | + | * Discuss the scenario for the next step's registration of prostates. | |
</div> | </div> | ||
| Line 22: | Line 24: | ||
<h1>Approach, Plan </h1> | <h1>Approach, Plan </h1> | ||
| − | Currently the algorithms have been implemented either in ITK or local C++ | + | Currently the algorithms have been implemented either in ITK or local C++ code. We'll discuss with Core 3 collaborators to put that into the software they are using to help with the segmentation work. |
</div> | </div> | ||
| Line 28: | Line 30: | ||
<h1>Progress</h1> | <h1>Progress</h1> | ||
| − | + | During the project week: | |
| + | * done: 3D Random Walk Segmentation Slicer3 command line module. | ||
| + | * done: [[Projects:ConformalFlatteningRegistration#Conformal_Flattening_Slicer3_module|Conformal Flattening]] ITK filter plugged in Slicer3 as a command line module. | ||
</div> | </div> | ||
Latest revision as of 13:50, 27 June 2008
Home < 2008 Summer Project Week:ProstateSegReg Return to Project Week Main Page |
Key Investigators
- Allen Tannenbaum, Yi Gao, Georgia Tech
- Gabor Fichtinger, Purang Abolmaesumi, Siddharth Vikal, David Gobbi, Queen's University
- Luis Ibanez, Kitware
Objective
- To integrate current segmentation algorithm into the software environment being used by Core 3 people.
- Discuss the scenario for the next step's registration of prostates.
Approach, Plan
Currently the algorithms have been implemented either in ITK or local C++ code. We'll discuss with Core 3 collaborators to put that into the software they are using to help with the segmentation work.
Progress
During the project week:
- done: 3D Random Walk Segmentation Slicer3 command line module.
- done: Conformal Flattening ITK filter plugged in Slicer3 as a command line module.