Difference between revisions of "2010 Winter Project Week SPECTRE 3DSlicer Integration"
| (7 intermediate revisions by the same user not shown) | |||
| Line 119: | Line 119: | ||
* c to configure | * c to configure | ||
* Set any missing values | * Set any missing values | ||
| − | **MIPAV should point to your installed version of mipav | + | **MIPAV should point to your installed version of mipav: SPECTRE_Plugin/mipav |
| − | + | **You can point the SPECTRE_ATLAS_TXT to the downloaded atlas: SPECTRE_Plugin/spectre_atlases/oasis-3.txt | |
| − | **You can point the SPECTRE_ATLAS_TXT to the downloaded atlas | + | **CMAKE_INSTALL_PREFIX to SPECTRE_Plugin/SPECTRE-install |
| − | |||
* c to configure | * c to configure | ||
** if it complains about not finding the INPUT_AXIAL_SUBSAMPLE file, c again or set it if you have a test image | ** if it complains about not finding the INPUT_AXIAL_SUBSAMPLE file, c again or set it if you have a test image | ||
| Line 128: | Line 127: | ||
* Build | * Build | ||
gmake | gmake | ||
| − | * If the build succeeds, there should be two important files | + | make install |
| + | * If the build succeeds, there should be two important files in SPECTRE_Plugin/SPECTRE-install/lib/Slicer3 | ||
SPECTRE.tcl | SPECTRE.tcl | ||
SPECTRE_Slicer.jar | SPECTRE_Slicer.jar | ||
| Line 134: | Line 134: | ||
* Under View->Application Settings go to the Module Settings page | * Under View->Application Settings go to the Module Settings page | ||
** add a directory pointing to the location of SPECTRE.tcl | ** add a directory pointing to the location of SPECTRE.tcl | ||
| − | * Restart Slicer3 and look for the new plugin | + | * Restart Slicer3 and look for the new plugin under Modules->Developer Tools |
====To run SPECTRE:==== | ====To run SPECTRE:==== | ||
| Line 149: | Line 149: | ||
====Trouble shooting==== | ====Trouble shooting==== | ||
| − | * | + | * If the SPECTRE building step fails with an error about not being able to find MIPAV, add the path to mipav to your PATH environment variable |
| − | + | * As of October 29/10 the SPECTRE cvs repository was not building, here's what I did to get it to build: | |
| − | ** you' | + | ** download the L-M fit package http://users.utu.fi/jaolho/ and unzip it in toads-cruise/releases/spectre/jistCRUISE/JIST |
| + | ** comment out all references to the missing class edu.jhu.ece.iacl.algorithms.graphics.utilities.GeodesicNeighborsOnSurface in the file jistCRUISE/JIST/src/edu/jhu/ece/iacl/algorithms/graphics/utilities/SurfaceMetrics.java | ||
| + | ** change the printout at line 184 of to print to err instead of out | ||
| + | toads-cruise/src/edu/jhu/ece/iacl/plugins/segmentation/skull_strip/MedicAlgorithmSPECTRE2009.java | ||
| + | * Use make, make clean, make install to rebuild without the extend.tcl script | ||
| + | * After compiling try running the tests: | ||
| + | cd Slicer3-ext/SPECTRE-build | ||
| + | ctest -R SPECTRE -VV | ||
| + | * Try running it manually, the following command should print out the XML description of the parameters with '''nothing''' before <?xml (otherwise Slicer won't recognise it as a plugin) | ||
| + | tclsh ./SPECTRE.tcl --xml | ||
| + | * If the XML is not printed out, you can try passing a bogus argument to see what the class path settings are (look for the path to MIPAV to verify that it's picking up the correct version) | ||
| + | tclsh ./SPECTRE.tcl --bogus | ||
| + | * Look in fileError.txt (standard error is written to this file) | ||
| + | ** if there's an error similar to:cli PARSE Error: Invalid ProcessingAlgorithm: edu.jhu.ece.iacl.plugins.segmentation.skull_strip.MedicAlgorithmSPECTRE2009 | ||
| + | ** then run MIPAV to set up some defaults (you may have to do this more than once if you're re-compiling). Instructions from Min: | ||
<pre> | <pre> | ||
1.)Close out of everything | 1.)Close out of everything | ||
| Line 167: | Line 181: | ||
8.)Try running it in Slicer again | 8.)Try running it in Slicer again | ||
</pre> | </pre> | ||
| − | |||
| − | |||
| − | |||
* If the SPECTRE module doesn't load into Slicer, exit and restart it with the --verbose-module-discovery command line option and look in the error log to see what was reported about the module discovery process for SPECTRE.tcl (the wrapper script for the .jar file) | * If the SPECTRE module doesn't load into Slicer, exit and restart it with the --verbose-module-discovery command line option and look in the error log to see what was reported about the module discovery process for SPECTRE.tcl (the wrapper script for the .jar file) | ||
| + | ** When tclsh SPECTRE.tcl is run with the --xml flag, nothing can be printed to standard output before <?xml or the module will not be recognised as a plugin | ||
| + | * The Mask output volume isn't loaded as a label map, you can reset it in the Volumes module, Info pane | ||
| + | **it also does not inherit the scalar type of the original volume. Cast the output volume to avoid problems with the EM Segmenter module | ||
| + | * The output volumes will sometimes have a flipped orientation, you can fix it in Slicer by: | ||
| + | ** In the Data module, right click on Scene and Add Transform | ||
| + | ** Drag the output volumes under the transform | ||
| + | ** Right click on the transform and click Edit Properties | ||
| + | ** Set element 1,1 to -1. | ||
===January 2010=== | ===January 2010=== | ||
Latest revision as of 15:36, 29 October 2010
Home < 2010 Winter Project Week SPECTRE 3DSlicer Integration
Key Investigators
- BWH: Nicole Aucoin
- JHU: Aaron Carass, Min Chen
- GE: Jim Miller
Objective
Integration of SPECTRE Skull Stripping software into the NAMIC Toolkit via the Slicer3 CLI framework.
Approach, Plan
Our approach: integration of software which will segment and provide coordinate reference frames for the Cerebrum (akin to the Talairach space), the Cerebellum (see reference below for K. Rehm), and for the Brainstem.
This will be achieved through integration of the Java Imaging Software Toolkit (JIST) into the Slicer3 framework and then testing and evaluation of JIST components for completing the necessary components to provide this system.
The main challenge is supporting Java command line executables in 3D Slicer.
Our plan is first to facilitate Java command line module discovery and execution, by adding support for the --xml argument to SPECTRE, and adding support for tcl wrapping scripts in 3DSlicer. Then integrate SPECTRE into the NAMIC toolkit by crafing a CMakeLists.txt file to allow it to be built easily. Create a NITRC project and a 3D Slicer extension that can be either downloaded or built on any platform.
Progress
A proof of concept sample Java HelloWorld application has been added to 3D Slicer.
Changes to the Command Line Module logic and Module Factory were made to support launching the tcl script with tclsh, for cross platform compatibility.
Changes made to the Java classes to support the --xml argument, Slicer recognises SPECTRE as a command line module.
Reading in the output images from SPECTRE back into Slicer works, to do: volume origin and win/level, stderr issue, marking some as label maps.
Updating instructions on how to run SPECTRE in Slicer to reflect current methods.
Presentations
Powerpoint presentation given by Jerry L. Prince on the status of the integration of SPECTRE as a module within Slicer, given at the 2010 Winter AHM in Utah.
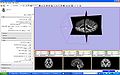
How to run SPECTRE with Slicer
October 2010
SPECTRE has been modified to work as an extension. This is still an intermediate stage in the integration process, the eventual end goal is to use the Extensions Wizard to download SPECTRE automatically.
The atlas can be found SPECTRE_ATLASES.zip
Build and install SPECTRE as an extension module in Slicer3
Currently the extension installation process is as follows (for linux and windows using cygwin):
- build Slicer3 from the source code in the SVN trunk
- Install MIPAV 5.0.0 or later
- set the install directory on linux to be Slicer3-lib/mipav. The default c:/Program Files/ location on windows should be fine.
- Download the extension file SPECTRE.s3ext.gz
- use gunzip to expand it
- put it in Slicer3/Extensions
- cd Slicer3/Scripts
- ./extend.tcl SPECTRE
- this will build the extension
- it checks for the required version of MIPAV and points to the .jar files there
- requires a Java compiler at least version 1.6 (to verify, you won't go wrong with the latest one)
- if you get a compiler error related to not finding a class, there's a possibility that the latest source code that you just checked out isn't quite in sync with the version of MIPAV or the CMake system, please contact Nicole Aucoin for help
- a new modification will be in Slicer3-ext/SPECTRE/toads-cruise/releases/spectre/CMakeLists.txt to tag the checkout, you can try adjusting the value of JIST_CVS_TAG
- The script will zip up the files needed to run the module as an extension, in Slicer3-ext/SPECTRE-install/lib/Slicer3/SPECTRE-cvs*.zip
- Start up Slicer3-build/Slicer3
- Under View->Application Settings go to the Module Settings frame
- Click on Add a preset button to add the path Slicer3-ext/SPECTRE-install/lib/Slicer3
- Restart Slicer3
- Look for the new module under Modules, Developer Tools, SPECTRE2009
Build and install SPECTRE as an extension module for a prebuilt binary of Slicer3
- Make a project directory
mkdir SPECTRE_Plugin cd SPECTRE_Plugin
- Install MIPAV 5.0.0 or later
- set the install directory on linux to be SPECTRE_Plugin/mipav. The default c:/Program Files/ location on windows should be fine.
- Download the atlas SPECTRE_ATLASES.zip and install in SPECTRE_Plugin/spectre_atlases
- Check out the CMake enabled source code
mkdir SPECTRE mkdir SPECTRE-build cd SPECTRE cvs -d :pserver:slicerjist:slicer@www.nitrc.org:/cvsroot/toads-cruise co toads-cruise/releases/spectre/
- Run ccmake to configure
cd ../SPECTRE-build ccmake ../SPECTRE/toads-cruise/releases/spectre/
- c to configure
- Set any missing values
- MIPAV should point to your installed version of mipav: SPECTRE_Plugin/mipav
- You can point the SPECTRE_ATLAS_TXT to the downloaded atlas: SPECTRE_Plugin/spectre_atlases/oasis-3.txt
- CMAKE_INSTALL_PREFIX to SPECTRE_Plugin/SPECTRE-install
- c to configure
- if it complains about not finding the INPUT_AXIAL_SUBSAMPLE file, c again or set it if you have a test image
- g to generate
- Build
gmake make install
- If the build succeeds, there should be two important files in SPECTRE_Plugin/SPECTRE-install/lib/Slicer3
SPECTRE.tcl SPECTRE_Slicer.jar
- Start up the Slicer3 binary
- Under View->Application Settings go to the Module Settings page
- add a directory pointing to the location of SPECTRE.tcl
- Restart Slicer3 and look for the new plugin under Modules->Developer Tools
To run SPECTRE:
- Download and unzip the altas from SPECTRE_ATLASES.zip
- Load an input volume, for example the first one from the atlas (spectre_atlases/OAS1_0001_MR1_original.hdr)
- Select that input volume in the SPECTRE GUI under the Volume drop down menu
- Click on the Atlas list folder and browse to your atlas file list (spectre_atlas/oasis-3.txt)
- Scroll down to the Output pane in the SPECTRE GUI and click on the drop down menus for the output volumes that you wish to reload into Slicer, for example, Stripped Volume, and select the Create New Volume option. You can also rename the volumes via the drop down menus.
- Click the Apply button
- Wait, the stripped brain is eventually loaded back into Slicer.
- It can take a few minutes to finish.
- If the "Completed with errors" message comes up, check the Error Log for the output from the module.
Trouble shooting
- If the SPECTRE building step fails with an error about not being able to find MIPAV, add the path to mipav to your PATH environment variable
- As of October 29/10 the SPECTRE cvs repository was not building, here's what I did to get it to build:
- download the L-M fit package http://users.utu.fi/jaolho/ and unzip it in toads-cruise/releases/spectre/jistCRUISE/JIST
- comment out all references to the missing class edu.jhu.ece.iacl.algorithms.graphics.utilities.GeodesicNeighborsOnSurface in the file jistCRUISE/JIST/src/edu/jhu/ece/iacl/algorithms/graphics/utilities/SurfaceMetrics.java
- change the printout at line 184 of to print to err instead of out
toads-cruise/src/edu/jhu/ece/iacl/plugins/segmentation/skull_strip/MedicAlgorithmSPECTRE2009.java
- Use make, make clean, make install to rebuild without the extend.tcl script
- After compiling try running the tests:
cd Slicer3-ext/SPECTRE-build ctest -R SPECTRE -VV
- Try running it manually, the following command should print out the XML description of the parameters with nothing before <?xml (otherwise Slicer won't recognise it as a plugin)
tclsh ./SPECTRE.tcl --xml
- If the XML is not printed out, you can try passing a bogus argument to see what the class path settings are (look for the path to MIPAV to verify that it's picking up the correct version)
tclsh ./SPECTRE.tcl --bogus
- Look in fileError.txt (standard error is written to this file)
- if there's an error similar to:cli PARSE Error: Invalid ProcessingAlgorithm: edu.jhu.ece.iacl.plugins.segmentation.skull_strip.MedicAlgorithmSPECTRE2009
- then run MIPAV to set up some defaults (you may have to do this more than once if you're re-compiling). Instructions from Min:
1.)Close out of everything 2.)Load up Mipav 3.)Go to the "Plugins" menu 4)If there is a "JIST" option then skip to 5.) otherwise do 4a-b.) 4a.)Select "Install Plugin" and install "Slicer3-ext/SPECTRE-install/lib/Slicer3/SPECTRE_Slicer.jar" (It might throw an error saying it can't be installed, that's okay) 4b.)Restart mipav 5.)Select Plugins, Click "JIST"->"JISTLayoutTool" 6.)If it asks you to pick a library directory, just pick an empty folder anywhere 7.)Close everything 8.)Try running it in Slicer again
- If the SPECTRE module doesn't load into Slicer, exit and restart it with the --verbose-module-discovery command line option and look in the error log to see what was reported about the module discovery process for SPECTRE.tcl (the wrapper script for the .jar file)
- When tclsh SPECTRE.tcl is run with the --xml flag, nothing can be printed to standard output before <?xml or the module will not be recognised as a plugin
- The Mask output volume isn't loaded as a label map, you can reset it in the Volumes module, Info pane
- it also does not inherit the scalar type of the original volume. Cast the output volume to avoid problems with the EM Segmenter module
- The output volumes will sometimes have a flipped orientation, you can fix it in Slicer by:
- In the Data module, right click on Scene and Add Transform
- Drag the output volumes under the transform
- Right click on the transform and click Edit Properties
- Set element 1,1 to -1.
January 2010
Linux
Updated instructions, based on the Windows process, to run SPECTRE with Slicer on a linux workstation (Jan 29/10):
- build Slicer3 from the source code in the SVN trunk
- install the SPECTRE classes and supporting classes in Slicer3/Applications/CLI/JAVA/SPECTRE/, so a plugins directory appears there that has all the supporting classes (ie the org tree) and the edu tree.
- Install MIPAV
- download installMIPAV.bin
- chmod u+x installMIPAV.bin
- sh ./installMIPAV.bin
- In the installer, Choose Install folder: Slicer3/Applications/CLI/Java/SPECTRE/mipav
- In the installer, Create links: none
- MIPAV comes with a JRE, to use that version of java: add Slicer3/Applications/CLI/Java/SPECTRE/mipav/jre/bin to the start of your path (Slicer will use it for the SPECTRE jars if you have the more recent version of SPECTRE.tcl)
- go to the Slicer3 source directory Slicer3\Applications\CLI and edit the file CMakeLists.txt
- remove the hash mark at the start of the line with Java on it (in the SET cli_modules command)
- in the Slicer3\Applications\CLI\Java directory, edit the file SPECTRE.tcl, and edit the following lines to point to your installation:
- pathToMipav to the full path to Slicer3/Applications/CLI/Java/SPECTRE/mipav
- pathToSPECTREPlugins to the full path to Slicer3/Applications/CLI/Java/SPECTRE/plugins
- Rebuild Slicer using Slicer3/Scripts/getbuildtest.tcl in order to install the helper script SPECTRE.tcl
- Start up Slicer
- check under the Modules menu, Developer Tools, and run the Java Example first. If the status message says "Completed with errors", you can restart Slicer with the --clear-module-cache argument and try it again.
- once it does run, SPECTRE 2009 is also categorised under Developer Tools, click on it to open it's GUI
- Load an input volume
- Select that input volume in the SPECTRE GUI under the Volume drop down menu (Slicer3/Application/CLI/Java/SPECTRE/mipav/plugins/atlases/spectre_atlas/OAS1_0001_MR1_original.hdr)
- Click on the Atlas list folder and browse to your atlas file list (Slicer3/Application/CLI/Java/SPECTRE//mipav/plugins/atlases/spectre_atlas/oasis-3_20081124.txt)
- Scroll down to the Output pane in the SPECTRE GUI and click on the drop down menus for the output volumes that you wish to reload into Slicer, for example, Stripped Volume, and select the Create New Volume option.
- Click the Apply button
- Wait, the stripped brain is eventually loaded back into Slicer.
Windows
At the project week, the integration was done on Nicole's Win32 laptop using these steps:
- build Slicer3 from the source code in the SVN trunk
- install the SPECTRE classes in c:\Documents and Settings\nicole\mipav\plugins, there should be a directory called edu there that holds the SPECTRE classes, as well as support libraries
- download and install MIPAV to C:\Program Files\mipav
- run MIPAV and under the Plugins menu, select Install Plugin.
- browse to "c:\Documents and Settings\nicole\mipav\plugins" and click on the arrow to move it the Selected class files window, then click Install Plugin(s)
- go to the Slicer3 source directory Slicer3\Applications\CLI and edit the file CMakeLists.txt
- remove the hash mark at the start of the line with Java on it (in the SET cli_modules command)
- in the Slicer3\Applications\CLI\Java directory, edit the file SPECTRE.tcl, and edit the following lines to point to your installation:
- set pathToMipavPlugins "c:/Program Files/mipav"
- lappend jarList {c:/Program Files/mipav}
- lappend jarList {c:/Documents and Settings/nicole/mipav/plugins}
- Rebuild Slicer using Slicer3/Scripts/getbuildtest.tcl in order to install the helper script SPECTRE.tcl
- Start up Slicer
- check under the Modules menu, Developer Tools, and run the Java Example first. If the status message says "Completed with errors", you can restart Slicer with the --clear-module-cache argument and try it again.
- once it does run, SPECTRE 2009 is also categorised under Developer Tools, click on it to open it's GUI
- Load an input volume
- Select that input volume in the SPECTRE GUI under the Volume drop down menu (c:/program files/mipav/plugins/atlases/spectre_atlas/OAS1_0001_MR1_original.hdr)
- Click on the Atlas list folder and browse to your atlas file list (c:/program files/mipav/plugins/atlases/spectre_atlas/oasis-3_20081124.txt)
- Scroll down to the Output pane in the SPECTRE GUI and click on the drop down menus for the output volumes that you wish to reload into Slicer, for example, Stripped Volume, and select the Create New Volume option.
- Click the Apply button
- Wait, the stripped brain is eventually loaded back into Slicer.
References
- K. Rehm's 2000 HBM abstract, 'Use of Cerebellar Landmarks To Define a Coordinate System and an Isolation Strategy'