Difference between revisions of "2011 Winter Project Week:SPECTRE Integration"
From NAMIC Wiki
| (8 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-SLC2011.png|[[2011_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2011.png|[[2011_Winter_Project_Week#Projects|Projects List]] | ||
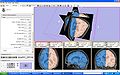
| + | Image:2011WinterProjectWeek-SPECTRE-3d.jpg|SPECTRE GUI and output | ||
| + | Image:2011WinterProjectWeek-SPECTRE-overlay.jpg|SPECTRE output brain mask as an overlay on input volume | ||
| + | Image:2011WinterProjectWeek-SPECTRE-volrend.jpg|SPECTRE skull stripped brain volume rendered | ||
</gallery> | </gallery> | ||
| Line 46: | Line 49: | ||
<li>Remove the requirement to have to run MIPAV before running the SPECTRE extension in Slicer. | <li>Remove the requirement to have to run MIPAV before running the SPECTRE extension in Slicer. | ||
<li>Set volume types in SPECTRE outputs | <li>Set volume types in SPECTRE outputs | ||
| + | <li>Find info on current state of batch jobs via clusters (for validation, not critical as they can run the java processes through the pipeline at JHU, but could be interesting) | ||
</ul> | </ul> | ||
| Line 54: | Line 58: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | During Project Week: | ||
| + | |||
| + | *Memory argument | ||
| + | **It will be difficult to add this argument to SPECTRE (SPECTRE won't use it, so adding it to the auto generated xml is tricky, and hard coding it in would be non optimal). | ||
| + | **For now, keep it as a command line module option that is stripped out before calling SPECTRE | ||
| + | **Checked in new SPECTRE.tcl.in to NITRC repository using the new argument | ||
| + | **To avoid the JVM crash when passing in a too large heap size, put in an if/else to call it with a small heap size if the argument is --xml | ||
| + | **Min hard coded the memory argument in JistCLI.java, working well in tests | ||
| + | |||
| + | *Python script | ||
| + | **Beta version written, talked to JC and he mentioned wanting to put native java support in Slicer4, so leave it for now | ||
| + | |||
| + | *Documentation | ||
| + | **expanded descriptions on parameters in xml | ||
| + | |||
| + | *Requirement to run MIPAV | ||
| + | ** Fix checked in to SPECTRE cvs, testing needed | ||
| + | |||
| + | * Y flip | ||
| + | ** SPECTRE reorients the input image to be axially aligned and isotropic, then it does output a realigned original image | ||
| + | ** Can Slicer use the output realigned original as the default bg/fg volume? | ||
| + | ** Min working on realigning the output to the input image | ||
| + | |||
| + | * Slicer Use Java Flag | ||
| + | ** Changes checked in to Slicer3 trunk, Slicer3_USE_JAVA set to OFF by default | ||
| + | ** Would be ideal to pass the cmake variable's value to extend.tcl so that it can be passed to the extensions modules via cmake. | ||
| + | ** Steve says that Slicer4 will use CMake to build extensions, but put the passing into extend.tcl so it can be ported, preliminary changes made and checked in to extend.tcl and SPECTRE's CMakeLists.txt file (will stop building if Slicer3_USE_JAVA is false). | ||
| + | |||
| + | Pre-project week: | ||
| + | <ul> | ||
| + | <li>Min checked in changes to the xml command line parameters to support label type volumes | ||
| + | <li>Memory | ||
<ul> | <ul> | ||
| + | <li>Command line argument name suggestions: -Xmx (too obscure), -memoryMax, -maximumMemory, -maxMemoryUsage | ||
| + | </ul> | ||
<li>Y flip | <li>Y flip | ||
<ul> | <ul> | ||
Latest revision as of 17:14, 14 January 2011
Home < 2011 Winter Project Week:SPECTRE IntegrationKey Investigators
- BWH: Nicole Aucoin
- JHU: Min Chen
Objective
To advance the integration of the Java based SPECTRE algorithm into Slicer3 (and Slicer4), to find validation data sets.
Approach, Plan
Our approach is to fine tune the integration for Slicer3, write a python wrapping script for Slicer4, fine tune the ACPC algorithm. The main challenge to this approach is coordinate system transforms.
Our plan for the project week is:
- Add a memory argument to the SPECTRE xml and use it to set Java heap sizes
- Fine tune the Java handling in Slicer3 to fail gracefully if Java is not available on a build machine
- Debug the ACPC algorithm in SPECTRE
- Add a transform output to SPECTRE
- Debug y flip on reading in output images into Slicer
- Find collaborators with segemented data sets for the validation portion of our project. Ron has suggested talking with:
- Sylvain Bioux
- Hans Johnson
- Write a python wrapper so we can integrate SPECTRE into Slicer4
- Work on documentation so that the manual steps are as clear as possible
- Remove the requirement to have to run MIPAV before running the SPECTRE extension in Slicer.
- Set volume types in SPECTRE outputs
- Find info on current state of batch jobs via clusters (for validation, not critical as they can run the java processes through the pipeline at JHU, but could be interesting)
Progress
During Project Week:
- Memory argument
- It will be difficult to add this argument to SPECTRE (SPECTRE won't use it, so adding it to the auto generated xml is tricky, and hard coding it in would be non optimal).
- For now, keep it as a command line module option that is stripped out before calling SPECTRE
- Checked in new SPECTRE.tcl.in to NITRC repository using the new argument
- To avoid the JVM crash when passing in a too large heap size, put in an if/else to call it with a small heap size if the argument is --xml
- Min hard coded the memory argument in JistCLI.java, working well in tests
- Python script
- Beta version written, talked to JC and he mentioned wanting to put native java support in Slicer4, so leave it for now
- Documentation
- expanded descriptions on parameters in xml
- Requirement to run MIPAV
- Fix checked in to SPECTRE cvs, testing needed
- Y flip
- SPECTRE reorients the input image to be axially aligned and isotropic, then it does output a realigned original image
- Can Slicer use the output realigned original as the default bg/fg volume?
- Min working on realigning the output to the input image
- Slicer Use Java Flag
- Changes checked in to Slicer3 trunk, Slicer3_USE_JAVA set to OFF by default
- Would be ideal to pass the cmake variable's value to extend.tcl so that it can be passed to the extensions modules via cmake.
- Steve says that Slicer4 will use CMake to build extensions, but put the passing into extend.tcl so it can be ported, preliminary changes made and checked in to extend.tcl and SPECTRE's CMakeLists.txt file (will stop building if Slicer3_USE_JAVA is false).
Pre-project week:
- Min checked in changes to the xml command line parameters to support label type volumes
- Memory
- Command line argument name suggestions: -Xmx (too obscure), -memoryMax, -maximumMemory, -maxMemoryUsage
- Y flip
- Atlas file OAS1_0001_MR1_original.hdr & .img loaded into Slicer using Add Data. Saved in .nhdr nrrd format, .nhdr contains:
type: short dimension: 3 space: left-posterior-superior sizes: 176 208 176 space directions: (1,0,0) (0,-1,0) (0,0,1) kinds: domain domain domain endian: little encoding: gzip space origin: (0,0,0)
- Loading the .nhdr file back into Slicer, volume aligns with .hdr/.img
- Turning off deleting intermediate files for SPECTRE: [$::slicer3::CommandLineModuleGUI_SPECTRE_2009 GetLogic] DeleteTemporaryFilesOff
- Running the algorithm, generating only stripped image. unu head on input image BJAIF_vtkMRMLScalarVolumeNodeB.nrrd:
type: short dimension: 3 space: left-posterior-superior sizes: 176 208 176 space directions: (1,0,0) (0,-1,0) (0,0,1) kinds: domain domain domain endian: little encoding: gzip space origin: (0,0,0)
- unu head on output stripped image BJAIF_vtkMRMLScalarVolumeNodeD.nrrd:
type: float dimension: 3 sizes: 176 208 176 endian: big encoding: raw kinds: space space space space: left-posterior-superior space origin: (-0.0,0.0,0.0) space directions: (1.0,0,0) (0,1.0,0) (0,0,1.0) thicknesses: NaN NaN 0.0
- Differences are the space directions, big instead of little endian, and the undefined thicknesses.
- Endian:
- Converting stripped image to big endian: unu save -f nrrd -en little -i /home/nicole/Slicer3nicole/BJAIF_vtkMRMLScalarVolumeNodeD.nrrd -o /home/nicole/Slicer3nicole/BJAIF_vtkMRMLScalarVolumeNodeD-littleEndian.nrrd
- and loading it back into Slicer, still offset
- Space directions
- Before this test, placing stripped output volume inside a transform with this matrix shifts it into alignment with the original image:
1 0 0 0 -1 0 0 0 1
but Dec 29/10, there's still an A-P flip.
- Before this test, placing stripped output volume inside a transform with this matrix shifts it into alignment with the original image:
- Atlas file OAS1_0001_MR1_original.hdr & .img loaded into Slicer using Add Data. Saved in .nhdr nrrd format, .nhdr contains:
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a
- ITK Module
- Slicer Module
- Built-in
- Extension -- commandline YES
- Extension -- loadable
References
- 2010 Summer Project Week
- 2010 Winter Project Week (has manual steps for integration)