Difference between revisions of "DBP:Harvard"
From NAMIC Wiki
| Line 32: | Line 32: | ||
{| cellpadding="5" | {| cellpadding="5" | ||
| − | ! width="120"| !! width=" | + | ! width="120"| !! width="300"| Title !! width="200" | Summary !! PIs |
|- | |- | ||
| [[Image:Shapeanalysis-UNC.jpg]] ||'''[[DBP:Harvard:Collaboration:UNC|Algorithms: Shape Analysis (PNL-UNC)]]''' || Structural shape analysis of the caudate nucleus and corpus callosum || <i>UNC</i>: Martin Styner, Isabelle Corouge<br><i>PNL</i>: Marek Kubicki, Sylvain Bouix, Marc Neithammer, James Levitt | | [[Image:Shapeanalysis-UNC.jpg]] ||'''[[DBP:Harvard:Collaboration:UNC|Algorithms: Shape Analysis (PNL-UNC)]]''' || Structural shape analysis of the caudate nucleus and corpus callosum || <i>UNC</i>: Martin Styner, Isabelle Corouge<br><i>PNL</i>: Marek Kubicki, Sylvain Bouix, Marc Neithammer, James Levitt | ||
|- | |- | ||
| − | | [[Image:Dti-UNC.jpg]] || '''Diffusion Tensor Imaging (PNL-UNC)''' || summary || <i>UNC</i>: Isabelle Corouge, Martin Styner<br><i>PNL:</i>Sylvain Bouix, Marc Niethammer, Marek Kubicki, Martha Shenton | + | | [[Image:Dti-UNC.jpg]] || '''[[DBP:Harvard:Collaboration:UNC|Algorithms: Diffusion Tensor Imaging (PNL-UNC)]]''' || summary || <i>UNC</i>: Isabelle Corouge, Martin Styner<br><i>PNL:</i>Sylvain Bouix, Marc Niethammer, Marek Kubicki, Martha Shenton |
|- | |- | ||
| [[Image:Dti-MIT.jpg]] || '''[[DBP:Harvard:Collaboration:MITDTI|Algorithms: Diffusion Tensor Imaging (PNL-MIT)]]''' || Automatic clustering of distinct fiber tracts, and diffusion measures along tracts of interest || <i>MIT</i>: Lauren O'Donnell, CF Westing, Raul San Jose<br><i>PNL:</i> Marek Kubicki, Sylvain Bouix, Marc Niethammer, Mark Dreusicke | | [[Image:Dti-MIT.jpg]] || '''[[DBP:Harvard:Collaboration:MITDTI|Algorithms: Diffusion Tensor Imaging (PNL-MIT)]]''' || Automatic clustering of distinct fiber tracts, and diffusion measures along tracts of interest || <i>MIT</i>: Lauren O'Donnell, CF Westing, Raul San Jose<br><i>PNL:</i> Marek Kubicki, Sylvain Bouix, Marc Niethammer, Mark Dreusicke | ||
Revision as of 19:00, 12 January 2007
Home < DBP:HarvardContents
IRB Information
In order to handle the data,
- You need IRB training (human subjects protection) and we need your proof--most PIs have taken the training and provided copy of the certificate (as a pre-requisite for the grant)--others handling data should do the same.
- Don't give the data to other people. If somebody wants the data, they should go and talk to Sylvain Bouix who will check if the data can be shared.
More guidelines will be discussed during the AHM next week.
Keep in mind that IRB training and certificates are mandatory and access to data is restricted to those with IRB approval.
- Note IRB must be updated annually in some institutions.
Data access
The description of the data can be found here.
- 2/2005: The legacy data has been converted to one single file format, nrrd, which contains information that we think is necessary for handling medical data. We will provide the NAMIC community documentation for the file format at the NAMIC meeting for both the medical and computer oriented public. We have a total of 188 morphological cases and 80 diffusion cases. The cases have a unique BIRN identifier. Information about diagnosis, data parameters and labels associated with Region of Interests (ROI) are on the BIRN site in the following Excel files:
- morph-NAMIC.xls : Gives diagnosis, age, gender as well as the ROIs drawn for each morphological case.
- morph-ROI-description.xls: Gives for each ROI set, the label corresponding to specific structures.
- diffusion-NAMIC.xls: Gives diagnosis, age, gender, ROIs drawn and when existing, the corresonding morphological data ID number.
- diffusion-ROI-description.xls: Gives or each ROI set, the label corresponding to specific structures.
- 1/2005: After discussion with Steve Pieper and Gordon Kindlman we are leaning towards the NIFTI-1 or nrrd format. NIFTI-1 has more info about orientation but is a bit cumbersome for multidimensional data (e.g. tensor). Nrrd lacks information about the world coordinate system. NOTE: orientation information has been added to NRRD, and to the "unu make" command-line tool that is the most convenient way to generate nrrd headers for a given raw datafile, so that now NRRD can losslessly encode all the orientation information found in the in the NIFTI-1 format.
- 12/2004: We are finalizing the collection of complete data sets. Formats vary across different data sets, genesis, signa, dicom, mrml... We need a single file format able that is simple and can handle orientation properly (i.e. not Analyze).
Ongoing Collaborations
This section is in the process of being updated
| Title | Summary | PIs | |
|---|---|---|---|
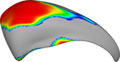 |
Algorithms: Shape Analysis (PNL-UNC) | Structural shape analysis of the caudate nucleus and corpus callosum | UNC: Martin Styner, Isabelle Corouge PNL: Marek Kubicki, Sylvain Bouix, Marc Neithammer, James Levitt |
 |
Algorithms: Diffusion Tensor Imaging (PNL-UNC) | summary | UNC: Isabelle Corouge, Martin Styner PNL:Sylvain Bouix, Marc Niethammer, Marek Kubicki, Martha Shenton |
 |
Algorithms: Diffusion Tensor Imaging (PNL-MIT) | Automatic clustering of distinct fiber tracts, and diffusion measures along tracts of interest | MIT: Lauren O'Donnell, CF Westing, Raul San Jose PNL: Marek Kubicki, Sylvain Bouix, Marc Niethammer, Mark Dreusicke |
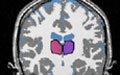 |
Algorithms: Anatomical Segmentation (PNL-MIT) | Brain tissue classification and parcellation of structures | MIT: Kilian Pohl, Sandy Wells, Eric Grimson PNL: Sylvain Bouix, Motoaki Nakamura, Min-Seong Koo, Martha Shenton |
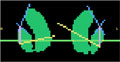 |
Algorithms: Rule-based Segmentation (PNL-GaTech) | Automatic segmentation of the striatum | GaTech: Ramsey Al-Hakim, Delphine Nain, Allen Tannenbaum PNL: Sylvain Bouix, James Levitt, Marc Niethammer, Martha Shenton |
| Algorithms: Tensor Based Statistics (PNL-Utah) | |||
| Engineering: Slicer Improvement and Testing | |||
| Training: Training Material and Expert Users Feedback |
Software Testing and Applications
- Software Test Bed: Contains 1) information about Slicer versions, and 2) data that can be used to verify software accuracy.
- Bugs: The ongoing list of technical issues for Slicer developers.
- New Tools: The "wish list" of research analysis tools, as well as progress on recent software additions.
Meeting Reports
- June 28, 2006: Meeting with Utah and UNC at the PNL during 2006 NA-MIC Project Week
- October 7, 2005, UNC visit at the Psychiatry NeuroImaging Laboratory
- Discuss two collaboration projects: Shape Analysis of Caudate Nucleus and Fiber Based Diffusion Analysis.
- 9/2005. DBP and Engineering Feedback Session
- Suggested implementation of tests for consistency of results (segmentation, registration, etc..) as the software evolves
- Gave some feedback on the slicer usage to other DBPs
- 5/2005 User training Seminar at Dartmouth
- Gave some presentations on useage of slicer by the lab
- Hands on help during the training sessions
- 3/2005 Visit from Ross Whitaker and Tom Fletcher from UofU.
- Presentation of non linear statistics for tensors by Tom Fletcher.
- Possible collaboration on comparing new and old anisotropy measures in the context of Schizophrenia.
- 2/2005 Utah AHM.
- prepared and presented the data.
- participated in the DTI engineering workshop.
- 12/2004 Software development meeting.
- Tina Kapur & Steve Pieper: Intro on the NAMIC project.
- Presentations of existing software from the different institutes:
- UNC, Martin Styner: Presentation of existing ITK tools created at UNC. Some interesting software for DTI processing.
- MIT, Lauren O'Donnell: Presentation of MIT algorithms. Polina Golland's shape analysis methods (classifier based on DT and statistical classifier). Kilian Pohl's Atlas Based EM segmentation which we are already using.
- Gtech, Delphine Nain: Presentation of GTech tools. Surface Evolution segmentation tool, Fast marching implementation in Slicer. Anisotropic image smoothing already in slicer. Future will be shape analysis (wavelets), stochastic curve evolution.
- UCLA: presentation of LONI, a pipeline building tool. Release date for beta testing is 5/2005.
- MGH, Dave Tuch: Presentation of the tools for Diffusion Imaging at MGH. Two major contributions: Q-ball, a high angular resolution imaging process; Sub parcellation of the thalamus based on DTI images.
- SPL: Presentation of the slicer by Steve Pieper and CF Westin. The DTI module is offering a number of very interesting properties. The schiz group is alpha testing it.
- Presentations on Software Development within the NAMIC framework:
- Luis Ibanez: Talk on Good Software Development Practices.
- Insight Toolkit (ITK) tutorial by Luis Ibanez. Highly templated modular library with many implemented image processing algorithms.
- Raul Estepar: How to integrate ITK classes in VTK.
- Steve Pieper: How to build and write code for the slicer.
- 11/2004 Introduced project to the Schizophrenia lab personnel
- RAs will be involved in collecting data and update site on patient recruitment
- Sylvain Bouix will be the point person for Core 3.1 @ Harvard
- 10/2004 Kickoff meeting