Projects:RegistrationLibrary:RegLib C06b
From NAMIC Wiki
Home < Projects:RegistrationLibrary:RegLib C06b
v3.6.1
Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.1  Registration Library Case #6B: RSNA 2011 DEMO Breast MRI Treatment Assessment
Registration Library Case #6B: RSNA 2011 DEMO Breast MRI Treatment Assessment
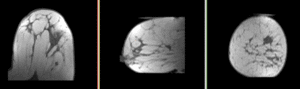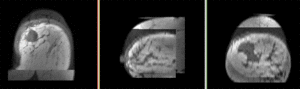
| this is the fixed reference image: PreRx Breast MRI with large tumor mass | 
|
this is the moving image, to be registered with the reference above: PostRx Breast MRI with tumor largely absent |
| fixed image/target pre Rx MRI |
moving image post Rx MRI |
Modules
- Slicer 3.6.1 recommended modules:
Objective / Background
We seek to align the post-treatment (PostRx) scan with the pre-treatment scan to compare local effects (left side only).
Keywords
MRI, breast cancer, intra-subject, treatment assessment, change detection, non-rigid registration
Download
- Data:
- Presets
Input Data
- reference/fixed : 0.44 x 0.44 x 5 mm , 784 x 784 x 30
- moving: 0.68 x 0.68 x 1.5 mm, 515 x 515 x 93
Methods
- Phase 1: affine alignment
- Go to the BRAINSfit module
- select Presets "Xf1_Affine" or set the parameters as given below:
- fixed image: "PreRx_left", moving image: "PostRx_left"
- Initialize with previous transform: select "Off"
- Initialize Transform Mode: check box for use MomentsAlign
- Registration Phases: check boxes for Include Rigid ..." and Include Affine registration phase
- Output: under Slicer Linear Transform, select new and rename to "Xf1_Affine" or similar
- Registration Parameters: this first phase is for initial alignment, we optimize/push for speed
- reduce "Number of Iterations" to 200
- reduce "Number of Samples" to 20,000
- leave rest at defaults
- Click Apply. Execution time ~ 10 seconds
- Phase 2: BSpline alignment
- Go to the BRAINSfit module
- select Presets "Xf2_BSpline1" or set the parameters as given below:
- fixed image: "PreRx_left", moving image: "PostRx_left"
- Initialize with previous transform: select "Xf1_Affine" from phase 1 above
- Initialize Transform Mode: check box for Off
- only check box for Include BSpline registration phase" , all other boxes off.
- Registration Parameters: set "Number of Samples" to 200,000 at least
- Output:
- Slicer BSpline Transform, select new and rename to "Xf2_BSpline" or similar
- Output Image Volume: select new and rename to "PostRx_left_Xf2" or similar
- Output Image Pixel Type: check box for "ushort"
- Registration Parameters:
- set "Number of Samples" to 100,000
- set Number of Grid Subdivisions to 7,7,5
- set Maximum B-Spline Displacement to 10 [mm]
- Click Apply. Execution time ~ 30 seconds
- Masking: check ROI box, Input Fixed Mask: "58_MRI-label", Input Moving Mask: "64_MRI-label" generated in Phase 3 above.
- Phase 3: Masking
- Before allowing nonrigid deformations we must mask the aliasing present:
- Go to the Editor module.
- Select 58_MRI as Master Volume, choose the threshold tool and threshold at intensity ~68 to separate from the background.
- Select the Brush tool and cut connections between the aliasing portions and the rest of the image
- Select Island Tool and remove aliasing portions
- Select Dilate tool to fill holes
- Save. Repeat for the 64_MRI volume
- Phase 4: BSpline alignment
- Go to the BRAINSfit module
- Initialize with previous transform: select "Xf2_Affine" from phase 2 above
- Output: under Slicer BSpline Transform, select new and rename to "Xf3_BSpline" or similar
- Output Image Volume: select new and rename to "64_MRI_Xf3" or similar
- Registration Parameters: set "Number of Samples" to 300,000 at least
- Number of Grid Subdivisions: 7,7,5
- Maximum B-Spline Displacement: set to 5 [mm]
- Masking: check ROI box, Input Fixed Mask: "58_MRI-label", Input Moving Mask: "64_MRI-label" generated in Phase 3 above.
- Click Apply
- Note to compare the different registrations: to see the Rigid or Affine registrations, drag the 64_MRI volume inside/onto the respective transform node. To see the BSpline registrations, select the "64_MRI_Xf3" volume as fore- or background.
- Extract left breast image of PreRx scan (ExtractSubvolumeROI module)
- Extract left breast image of PostRx scan (ExtractSubvolumeROI module)
- run MRI Bias field inhomogeneity correction on PreRx scan (MRI Bias Field Correction module)
- run affine registration (Robust Multiresolution Affine module)
- Fixed Image: PreRx_left_BiasCorr
- Moving Image: PostRx_left
- Resample Image: none
- Output transform: Create new linear transform, rename to: Xform_Aff0_MRes
- Fixed Image Mask: none
- Step Size (voxels):5
- Evaluate quality of Affine registration: drag PostRx_left inside the abovecreated Xform node (Data module)
- run Bspline non-rigid registration (Fast Deformable BSpline registration module)
- Iterations: 50
- Grid Size: 5
- Histogram Bins: 100
- Spatial Samples: 80000
- Constrain Deformation: no
- Initial Transform: XForm_Aff0_MRes
- Fixed Image: PreRx_left_BiasCorr
- Moving Image: PostRx_left
- Output Transform: Create New BSpline Transform, rename to: Xform_BSpline1_Aff0Init
- Output Volume: Create New Volume, rename to: PostRx_left_BSpline1
- Apply.
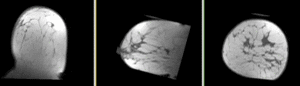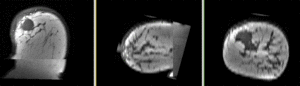
Registration Results
Discussion: Registration Challenges
- soft tissue deformations during image acquisition cause large differences in appearance
- the large tumor recession represents a significant pre/post difference in image content that will influence unmasked intensity-driven registration, which becomes a problem for the non-rigid portion of registration, particularly at higher DOF, because the registration will try to "recreate" the tumor area from the postRx image in order to match the content.
- contrast enhancement and pathology and treatment changes cause additional differences in image content
- the surface coils used cause strong differences in intensity inhomogeneity.
- we have strongly anisotropic voxel sizes with much less through-plane resolution
- resolution and FOV change between the two scans
Discussion: Key Strategies
- because of the strong changes in shape and position, we break the problem down and register each breast separately.
- we perform a bias-field correction on both images before registration
- we use the Multires version of RegisterImages for an initial affine alignment
- the nonlinear portion is then addressed with a BSpline or DiffeomorphicDemons algorithm
- because accuracy is more important than speed here, we increase the sampling rate (i.e. the number of points sampled for the BSpline registration)