Difference between revisions of "Projects:RegistrationLibrary:RegLib C17"
From NAMIC Wiki
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| Line 17: | Line 17: | ||
=== Modules === | === Modules === | ||
| − | *[ | + | *[https://www.slicer.org/wiki/Modules:BRAINSFit '''BRAINSfit'''] |
| − | *for mask generation: ''Draw'' tool in the [ | + | *for mask generation: ''Draw'' tool in the [https://www.slicer.org/wiki/Modules:Editor-Documentation-3.6 Editor] and/or [https://www.slicer.org/wiki/Modules:FastMarchingSegmentation-Documentation-3.6 Fast-Marching Segmentation], |
=== Keywords === | === Keywords === | ||
| Line 47: | Line 47: | ||
=== Procedures=== | === Procedures=== | ||
*'''Phase 1''': obtain masks for kidney ROI | *'''Phase 1''': obtain masks for kidney ROI | ||
| − | #Use the ''Draw'' tool in the [ | + | #Use the ''Draw'' tool in the [https://www.slicer.org/wiki/Modules:Editor-Documentation-3.6 Editor] and/or use the [https://www.slicer.org/wiki/Modules:FastMarchingSegmentation-Documentation-3.6 Fast-Marching Segmentation] module to obtain segmentations of the kidney in both CT and MR image. In the example dataset, you can find those segementations as files "Probe1CT_seg.nrrd" and "MRl_seg.nrrd" |
| − | #See the [ | + | #See the [https://www.slicer.org/wiki/Slicer_3.6:Training tutorials] for the Editor and and segmentation methods for details on how to obtain an efficient segmentation. |
*'''Phase 2''': obtain initial manual alignment | *'''Phase 2''': obtain initial manual alignment | ||
| − | **Details in the [ | + | **Details in the [https://www.slicer.org/wiki/Slicer3.6:Training manual registration tutorial] and also in the [https://www.slicer.org/wiki/Slicer-3-6-FAQ#How_do_I_initialize.2Falign_images_with_very_different_orientations_and_no_overlap.3F Slicer FAQ] |
| − | #go to the [ | + | #go to the [https://www.slicer.org/wiki/Modules:Data-Documentation-3.6 Data module] |
#right click on the "Scene" mode and select ''Insert New Transform''. In the MRML edit field below, rename it to "Xf0_manual" or similar | #right click on the "Scene" mode and select ''Insert New Transform''. In the MRML edit field below, rename it to "Xf0_manual" or similar | ||
#In the ''Data'' module, drag the "MRI" image volume inside the"Xf0_manual" node | #In the ''Data'' module, drag the "MRI" image volume inside the"Xf0_manual" node | ||
#Select the views so that the MR and CT volumes are displayed in the slice views | #Select the views so that the MR and CT volumes are displayed in the slice views | ||
| − | #Go to the [ | + | #Go to the [https://www.slicer.org/wiki/Modules:Transforms-Documentation-3.6 Transforms module] and adjust the translation and rotation sliders to adjust the current position. To get a finer degree of control, enter smaller numbers for the translation limits and enter rotation angles numerically in increments of a few degrees at a time |
*'''Phase 3''': automated rigid/affine alignment | *'''Phase 3''': automated rigid/affine alignment | ||
| − | #Go to the [ | + | #Go to the [https://www.slicer.org/wiki/Modules:BRAINSFit '''BRAINSfit'''] module |
##select Presets "Xf1_Rigid" or "Xf2_Affine" or set the parameters as given below: | ##select Presets "Xf1_Rigid" or "Xf2_Affine" or set the parameters as given below: | ||
##fixed image: "Probe1CT", moving image: "MRI" | ##fixed image: "Probe1CT", moving image: "MRI" | ||
| Line 67: | Line 67: | ||
##Mask Processing: check ''ROI'' box, ''Input Fixed Mask'' select CT segmentation created in Phase 1 above. For ''Input Moving Mask'' select MRI segmentation created in Phase 1 above. | ##Mask Processing: check ''ROI'' box, ''Input Fixed Mask'' select CT segmentation created in Phase 1 above. For ''Input Moving Mask'' select MRI segmentation created in Phase 1 above. | ||
##Click Apply | ##Click Apply | ||
| − | #return to the [ | + | #return to the [https://www.slicer.org/wiki/Modules:Data-Documentation-3.6 Data module] and drag the MRI inside/outside the different registration transforms to compare the alignment |
#to obtain a resampled volume: move MRI inside Xform of choice and then right-click on the volume and select ''Harden Transforms''. Save MRI under new name. | #to obtain a resampled volume: move MRI inside Xform of choice and then right-click on the volume and select ''Harden Transforms''. Save MRI under new name. | ||
Latest revision as of 17:36, 10 July 2017
Home < Projects:RegistrationLibrary:RegLib C17Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
Contents
v3.6.3  Slicer Registration Library Case #17: Kidney pre-op MR to intra-op CT
Slicer Registration Library Case #17: Kidney pre-op MR to intra-op CT
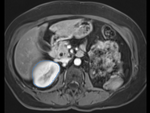
Input

|
|
|
| fixed image/target: intra-operative CT | moving image: per-operative MRI |
Modules
- BRAINSfit
- for mask generation: Draw tool in the Editor and/or Fast-Marching Segmentation,
Keywords
CT, MRI, abdominal, image-guided therapy, IGT, tumor ablation
Input Data
- fixed: intra-operative CT, oblique, 0.58 x 0.58 x 3 mm voxel size, dimensions 512 x 512 x 20
- moving: opre-operative MRI lique, 1.2 x 1.2 x 4 mm voxel size, dimensions 320 x 240 x 64
Download
- Data
- Presets
Registration Challenges
- intra-operative CT is acquired with limited FOV and very oblique position
- intra-operative CT contains artifacts from surgical needles
- both datasets have strong voxel anisotropy
- the region of interest (kidney) is small compared to the FOV, i.e. there is substantial "distracting" image content
Key Strategies
- manual alignment to obtain initial pose
- rough segmentation of both kidneys to mask "distracting" image content
- gradual increase in registration DOF.
Procedures
- Phase 1: obtain masks for kidney ROI
- Use the Draw tool in the Editor and/or use the Fast-Marching Segmentation module to obtain segmentations of the kidney in both CT and MR image. In the example dataset, you can find those segementations as files "Probe1CT_seg.nrrd" and "MRl_seg.nrrd"
- See the tutorials for the Editor and and segmentation methods for details on how to obtain an efficient segmentation.
- Phase 2: obtain initial manual alignment
- Details in the manual registration tutorial and also in the Slicer FAQ
- go to the Data module
- right click on the "Scene" mode and select Insert New Transform. In the MRML edit field below, rename it to "Xf0_manual" or similar
- In the Data module, drag the "MRI" image volume inside the"Xf0_manual" node
- Select the views so that the MR and CT volumes are displayed in the slice views
- Go to the Transforms module and adjust the translation and rotation sliders to adjust the current position. To get a finer degree of control, enter smaller numbers for the translation limits and enter rotation angles numerically in increments of a few degrees at a time
- Phase 3: automated rigid/affine alignment
- Go to the BRAINSfit module
- select Presets "Xf1_Rigid" or "Xf2_Affine" or set the parameters as given below:
- fixed image: "Probe1CT", moving image: "MRI"
- Initialize with previous transform: select "Xf0_manual" from Phase 1 above
- Initialize Transform Mode: leave at default = OFF
- check Include Rigid registration Phase box. For affine also check ScaleVersor3D and Affine
- Output: under Slicer Linear Transform, select new and rename to "Xf1_Rigid" or "Xf2_Affine" or similar
- Registration Parameters: set "Number of Samples" to 100,000 at least
- Mask Processing: check ROI box, Input Fixed Mask select CT segmentation created in Phase 1 above. For Input Moving Mask select MRI segmentation created in Phase 1 above.
- Click Apply
- return to the Data module and drag the MRI inside/outside the different registration transforms to compare the alignment
- to obtain a resampled volume: move MRI inside Xform of choice and then right-click on the volume and select Harden Transforms. Save MRI under new name.
Registration Results
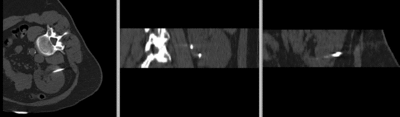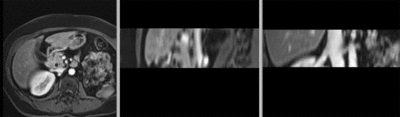 original unregistered
original unregistered
 after manual initial alignment
after manual initial alignment
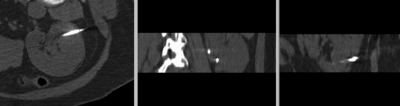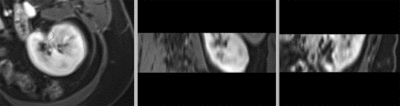 after rigid registration
after rigid registration
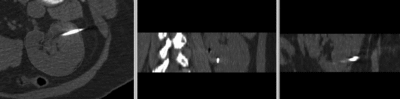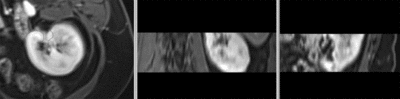 after affine registration
after affine registration