Difference between revisions of "Projects:RegistrationLibrary:RegLib C33"
From NAMIC Wiki
| Line 50: | Line 50: | ||
=== Procedures === | === Procedures === | ||
| − | *'''Phase I: | + | === Procedures === |
| − | # | + | *'''Phase I: Preprocessing: Build DWI mask + baseline''' |
| − | + | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/DiffusionWeightedMasking ''Modules:Diffusion:DiffusionWeightedImages:DiffusionWeightedVolumeMasking'' ] module | |
| − | + | ##''Input DWI Volume'': "DWI" | |
| − | + | ##''Output Baseline Volume'': ''Create New Volume'', rename to "DWI_baseline" | |
| − | ## | + | ##''Output Threshold Mask'': ''Create New Volume'', rename to "DWI_mask" |
| − | ## | + | ##Leave other settings at default; click ''Apply'' |
| − | ## | + | *'''Phase II: Preprocessing: Convert DWI -> DTI''' |
| − | + | #open [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/DiffusionTensorEstimation "Diffusion Tensor Estimation" module] (menu: Diffusion:DiffusionWeightedImages: DiffusionTensorEstimation) | |
| − | + | ##''Input DWI Volume'': DWI_iso, | |
| − | # | + | ##''Output DTI Volume'': create new, rename to "DTI_iso" |
| − | *'''Phase | + | ##''Output Baseline Volume'': create new, rename to "DWI_iso_baseline" |
| − | # | + | #Click: Apply |
| − | + | *'''Phase III: register FLAIR to T1Gd''' | |
| − | #Input DWI Volume: | + | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module] |
| − | #Output Baseline: create new, rename to " | + | ##''Fixed Image Volume'': FLAIR |
| − | # | + | ##''Moving Image Volume'': T1 |
| − | |||
| − | |||
| − | *'''Phase | ||
| − | #open Registration | ||
| − | ## | ||
| − | |||
| − | |||
| − | ## | ||
##Output Settings: | ##Output Settings: | ||
| − | ### | + | ###''Slicer BSpline Transform": none |
| − | ### | + | ###''Slicer Linear Transform'': create new transform, rename to "Xf1_FLAIR-T1Gd_Affine" |
| − | ##Registration Parameters: | + | ###''Output Image Volume'': create new volume, rename to "FLAIR_Xf1" |
| − | ## | + | ##''Registration Phases'': check boxes for ''Affine'' only |
| + | ##''Main Parameters'': | ||
| + | ###''Number Of Samples'': 200,000 | ||
| + | ##''Mask Option'': select ''ROIAUTO'' button | ||
| + | ###''(ROIAUTO) Output fixed mask'': create new volume | ||
| + | ###''(ROIAUTO) Output moving mask'': create new volume | ||
##Leave all other settings at default | ##Leave all other settings at default | ||
| − | ##click: Apply | + | ##click: Apply |
| − | + | *'''Phase IV: register T1 to T1Gd''' | |
| − | + | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module] | |
| − | *'''Phase | + | ##''Fixed Image Volume'': T1Gd |
| − | # | + | ##''Moving Image Volume'': T1 |
| − | ## | + | ##Output Settings: |
| − | ## | + | ###''Slicer BSpline Transform": none |
| − | ## | + | ###''Slicer Linear Transform'': create new transform, rename to "Xf2_T1-T1Gd_Affine" |
| − | ## | + | ###''Output Image Volume'': create new volume, rename to "T1_Xf2" |
| − | ## | + | ##''Registration Phases'': check boxes for ''Affine'' only |
| − | ## | + | ##''Main Parameters'': |
| − | ## | + | ###''Number Of Samples'': 200,000 |
| − | ## | + | ##''Mask Option'': select ''ROIAUTO'' button |
| − | # | + | ###''(ROIAUTO) Output fixed mask'': create new volume |
| − | # | + | ###''(ROIAUTO) Output moving mask'': create new volume |
| − | # | + | ##Leave all other settings at default |
| − | *'''Phase | + | ##click: Apply |
| − | #open Registration : '' | + | *'''Phase V: Register DTI (masked)''' |
| − | ## | + | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module] |
| − | ### | + | ##''Fixed Image Volume'': FLAIR_Xf1 |
| − | ### | + | ##''Moving Image Volume'': DWI_baseline |
| − | ## | + | ##Output Settings: |
| − | ## | + | ###''Slicer BSpline Transform": create new transform, rename to "Xf3_DTI-FLAIR" |
| − | ## | + | ###''Slicer Linear Transform'': none |
| − | ## | + | ###''Output Image Volume'': create new volume, rename to DWI_baseline_Xf2 |
| − | ## | + | ##''Regstration Phases'': check boxes for ''Rigid'', ''Affine'' and ''BSpline'' |
| − | ## | + | ##''Main Parameters'': |
| + | ###''Number Of Samples'': 300,000 | ||
| + | ###''B-Spline Grid Size'': 7,7,5 | ||
| + | ##''Mask Option'': select ''ROI'' button | ||
| + | ###''ROI Masking input fixed'': select " "FLAIR_mask_ROIauto" created in phase III above | ||
| + | ###''ROI Masking input moving'': select "DWI_mask" created in phase I above | ||
| + | ##Leave all other settings at default | ||
| + | ##click: Apply; runtime 1-2 min (MacPro QuadCore 2.4GHz) | ||
*'''Phase V: Resample DTI''' | *'''Phase V: Resample DTI''' | ||
| − | #Open the ''Resample DTI Volume'' module ( | + | #Open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/ResampleDTI ''Resample DTI Volume'' module] (under ''All Modules'' menu; note this is distinct from the ResampleScalarVectorDWIVolume used above) |
| − | ##Input Volume: | + | ##''Input Volume'': DTI |
| − | ##Output Volume | + | ##''Output Volume'': create new DTI Volume, rename to ''DTI_Xf3'' |
| − | ##Reference Volume | + | ##''Reference Volume'': T1Gd |
| − | ##Transform | + | ##''Transform Node'': select "Xf3_DTI-FLAIR" created above |
| − | ##check box: '' | + | ##check box: ''displacement'' |
| − | # | + | #leave all other settings at defaults |
| − | + | #Click Apply; runtime ~ 3 min. | |
| − | + | #set T1 or FLAIR as background and the new ''DTI_Xf2'' volume as foreground | |
| − | + | #Move fade slider to see DTI overlay onto the structural image | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | # | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Registration Results=== | === Registration Results=== | ||
Revision as of 19:42, 16 May 2012
Home < Projects:RegistrationLibrary:RegLib C33Back to ARRA main page
Back to Registration main page
Back to Registration Use-case Inventory
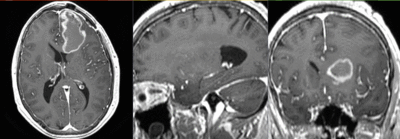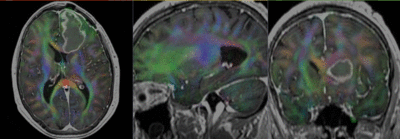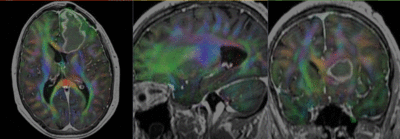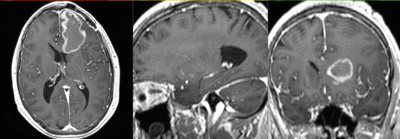
v3.6.1  Slicer Registration Library Exampe #33: Diffusion Weighted Image Volume: align with structural reference MRI
Slicer Registration Library Exampe #33: Diffusion Weighted Image Volume: align with structural reference MRI
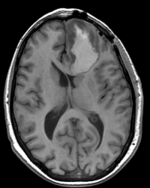
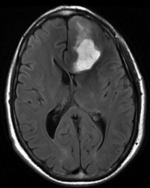
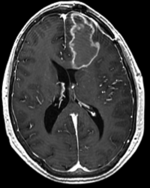
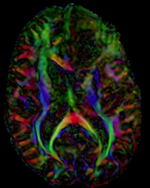
Input
|
|
|
|
|
|
| fixed image/target T1 |
moving image FLAIR |
moving image 2a T1Gd |
moving image 2b DTI tensor |
Modules
- Slicer 3.6.1 recommended modules: BrainsFit
Objective / Background
This is a typical example of DTI processing. Goal is to align the DTI image with a structural scan that provides accuracte anatomical reference. The DTI contains acquisition-related distortion and insufficient contrast to discern anatomical detail. For treatment planning and evaluation, location of functionally critical fiber tracts relative to the pathology is sought.
Keywords
MRI, brain, head, intra-subject, DTI, DWI
Input Data
- reference/fixed : T1 axial, 0.5 x 0.5 x 1 , 512 x 512 x 176
- moving : FLAIR axial
- moving : T1Gd axial
- moving: Tensor data of DWI volume, 32 directions, 2.6 mm slice thickness
Overall Strategy
- The best target for registering the DWI in terms of contrast is the FLAIR
- however the FLAIR is low resolution. The T1 is very high in-plane resolution, which causes memory issues when trying to resample the DTI to that resolution. So we choose the T1Gd as the reference space and resolution (1x1x1 mm).
- Thus the main strategy is:
- Obtain mask and baseline from DWI
- Compute DTI from DWI
- Register FLAIR and T1 to T1Gd (affine only) with masking (Affine)
- Register DWI_baseline to the resampled FLAIR (affine+nonrigid) (masking)
- Resample the DTI with above transform and the T1Gd as reference
For an alternative approach using Slicer 3.6 see here
Procedures
Procedures
- Phase I: Preprocessing: Build DWI mask + baseline
- open the Modules:Diffusion:DiffusionWeightedImages:DiffusionWeightedVolumeMasking module
- Input DWI Volume: "DWI"
- Output Baseline Volume: Create New Volume, rename to "DWI_baseline"
- Output Threshold Mask: Create New Volume, rename to "DWI_mask"
- Leave other settings at default; click Apply
- Phase II: Preprocessing: Convert DWI -> DTI
- open "Diffusion Tensor Estimation" module (menu: Diffusion:DiffusionWeightedImages: DiffusionTensorEstimation)
- Input DWI Volume: DWI_iso,
- Output DTI Volume: create new, rename to "DTI_iso"
- Output Baseline Volume: create new, rename to "DWI_iso_baseline"
- Click: Apply
- Phase III: register FLAIR to T1Gd
- open the General Registration (BRAINS) module
- Fixed Image Volume: FLAIR
- Moving Image Volume: T1
- Output Settings:
- Slicer BSpline Transform": none
- Slicer Linear Transform: create new transform, rename to "Xf1_FLAIR-T1Gd_Affine"
- Output Image Volume: create new volume, rename to "FLAIR_Xf1"
- Registration Phases: check boxes for Affine only
- Main Parameters:
- Number Of Samples: 200,000
- Mask Option: select ROIAUTO button
- (ROIAUTO) Output fixed mask: create new volume
- (ROIAUTO) Output moving mask: create new volume
- Leave all other settings at default
- click: Apply
- Phase IV: register T1 to T1Gd
- open the General Registration (BRAINS) module
- Fixed Image Volume: T1Gd
- Moving Image Volume: T1
- Output Settings:
- Slicer BSpline Transform": none
- Slicer Linear Transform: create new transform, rename to "Xf2_T1-T1Gd_Affine"
- Output Image Volume: create new volume, rename to "T1_Xf2"
- Registration Phases: check boxes for Affine only
- Main Parameters:
- Number Of Samples: 200,000
- Mask Option: select ROIAUTO button
- (ROIAUTO) Output fixed mask: create new volume
- (ROIAUTO) Output moving mask: create new volume
- Leave all other settings at default
- click: Apply
- Phase V: Register DTI (masked)
- open the General Registration (BRAINS) module
- Fixed Image Volume: FLAIR_Xf1
- Moving Image Volume: DWI_baseline
- Output Settings:
- Slicer BSpline Transform": create new transform, rename to "Xf3_DTI-FLAIR"
- Slicer Linear Transform: none
- Output Image Volume: create new volume, rename to DWI_baseline_Xf2
- Regstration Phases: check boxes for Rigid, Affine and BSpline
- Main Parameters:
- Number Of Samples: 300,000
- B-Spline Grid Size: 7,7,5
- Mask Option: select ROI button
- ROI Masking input fixed: select " "FLAIR_mask_ROIauto" created in phase III above
- ROI Masking input moving: select "DWI_mask" created in phase I above
- Leave all other settings at default
- click: Apply; runtime 1-2 min (MacPro QuadCore 2.4GHz)
- Phase V: Resample DTI
- Open the Resample DTI Volume module (under All Modules menu; note this is distinct from the ResampleScalarVectorDWIVolume used above)
- Input Volume: DTI
- Output Volume: create new DTI Volume, rename to DTI_Xf3
- Reference Volume: T1Gd
- Transform Node: select "Xf3_DTI-FLAIR" created above
- check box: displacement
- leave all other settings at defaults
- Click Apply; runtime ~ 3 min.
- set T1 or FLAIR as background and the new DTI_Xf2 volume as foreground
- Move fade slider to see DTI overlay onto the structural image
Registration Results
Download
- Data
- RegLib_C33_Data DWI, DTI, T1, FLAIR, Presets & solution transforms (NRRD files, zip file 174 MB)
- RegLib_C33_Data_noDWI subset of the above, contains DTI but w/o DWI and resampled DTI (NRRD files, zip file 86 MB)
- RegLib_C33_Data Transforms & Presets only contains only result transforms and parameter presets (NRRD files, zip file 30 kB)
- Presets
Discussion: Registration Challenges
- The DTI contains acquisition-related distortions (commonly EPI acquisitions) that can make automated registration difficult.
Discussion: Key Strategies
Acknowledgments
This case is the same data as case 40 from the fMRI neurosurgery data set on central.xnat.org