4D DTI analysis via longitudinal modeling of tract diffusivities
Key Investigators
Anuja Sharma, Bo Wang, Andrei Irimia, Micah Chambers, Jack Van Horn, Marcel Prastawa, Guido Gerig
Project Description
Objective
- Preliminary longitudinal evaluation of white matter diffusion changes along specific tracts in TBI subjects.
- Collaborate with the UCLA team to evaluate results and discuss improvements.
Approach, Plan
- Patient specific co-registration of acute and chronic DTI images.
- ROI based white matter tract extraction
- Assembling diffusion properties along tract in a space normalized fashion via a consistent mapping of tract geometry across timepoints.
- Evaluation of diffusion changes in white matter properties
Progress
- Preliminary workflow is ready. Details are provided in the following sections.
- Had collaborative meetings with the UCLA TBI team for discussion of current progress and future algorithm ideas. In particular, we focused on methodologies to quantify white matter changes along time in TBI patients.
- Resolved some TBI data issues with the help of Andrei Irimia (dicom conversion).
- Discussed possible registration options for DTI data in the context of TBI with Martin Styner (DTI Reg, DTI AtlasBuilder, ANTS).
- Got an insight into Slicer capabilities applicable to TBI from Ron.
- Attended the TBI DBP Meet.
Preliminary workflow for longitudinal evaluation of white matter changes in TBI
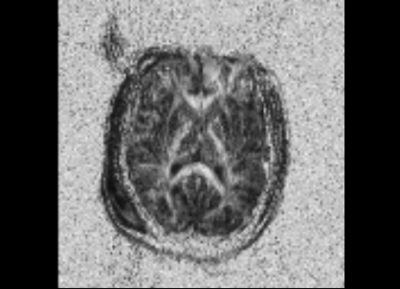
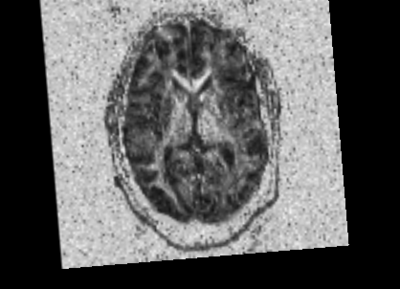
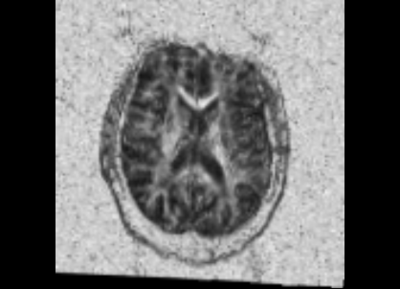
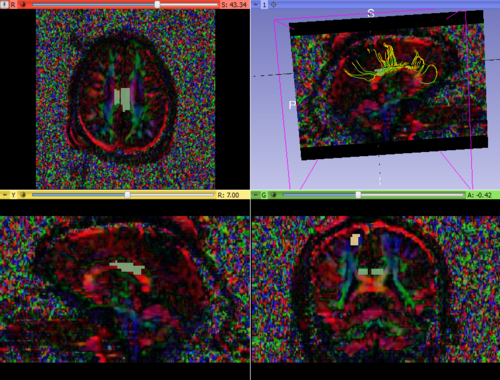
We started with the Dicom volumes for both the timepoints for a single TBI patient (mild TBI, id 16) and converted these to DTI nrrd volumes. Using the DTI volumes, we computed the respective FA volumes as well. For registration, we tried a few methods (DTI Reg utility, FA based registration-affine/Bspline etc). We used acute as fixed and chronic as the moving image. Given this was a proof of concept, rigid, followed by affine on FA images worked decently. We also computed mask volumes to drive the registration better (given all the DTI noise, artifacts and lesion related irregularities). However, the one that worked the best for now was without any specific masks. The linear transform obtained was used to resample the DTI chronic volume (in Slicer).
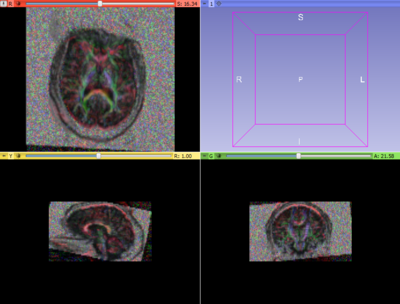
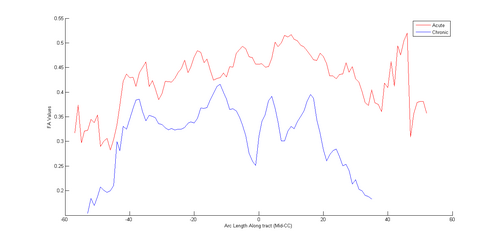
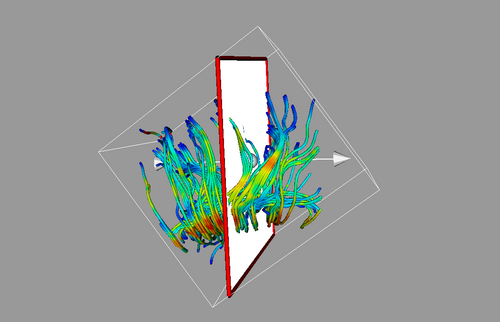
Once both time points were in the same space, we extracted ROIs in one of the images (either acute or resampled chronic). We tried out mid-CC for this study. This common ROI label map was then used for tractography in the acute and resampled volumes. The fiber tracts were then cleaned up, arc length parametrized and their FA profiles were compared.
In the future, we would like to improve the complete workflow wrt registration, ROI extraction and tractography. We also want to try out more cases, more regions of the brain and see the overall pattern of white matter changes in TBI.