Difference between revisions of "Algorithm:GATech"
| (110 intermediate revisions by 9 users not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[Algorithm:Main|NA-MIC Algorithms]] | + | Back to [[Algorithm:Main|NA-MIC Algorithms]] |
__NOTOC__ | __NOTOC__ | ||
| − | = Overview of | + | = Overview of Boston University Algorithms (PI: Allen Tannenbaum) = |
| − | At | + | At Boston University, we are broadly interested in a range of mathematical image analysis algorithms for segmentation, registration, diffusion-weighted MRI analysis, and statistical analysis. For many applications, we cast the problem in an energy minimization framework wherein we define a partial differential equation whose numeric solution corresponds to the desired algorithmic outcome. The following are many examples of PDE techniques applied to medical image analysis. |
| − | = | + | = Boston University Projects = |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {| cellpadding="10" style="text-align:left;" | |
| + | |||
| + | | | [[Image:MultiScaleHippoSegmentationHausdorf.png|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:MultiScaleShapeSegmentation|Multi-scale Shape Representation and Segmentation With Applications to Radiotherapy]] == | ||
| + | |||
| + | We present in this work a multiscale representation for shapes with arbitrary topology, and a method to segment the target organ/tissue from medical images having very low contrast with respect to surrounding regions using multiscale shape information and local image features. In a number of previous papers, shape knowledge was incorporated by first constructing a shape space from training data, and then constraining the segmentation process to be within the resulting shape space. However, such an approach has certain limitations including the fact that small scale shape variances may be overwhelmed by those on larger scale, and therefore the local shape information is lost. In this work, first we handle this problem by providing a multiscale shape representation using the wavelet transform. Consequently, the shape variances captured by the statistical learning step are also represented at various scales. In doing so, not only is the diversity of shape enriched, but also small scale changes are nicely captured. [[Projects:MultiScaleShapeSegmentation|More...]] | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:GT-SPD-img1.png|200px|]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:SoftPlaqueDetection|Soft Plaque Detection in CTA Imagery]] == | ||
| + | |||
| + | The ability to detect and measure non-calcified plaques (also known as soft plaques) may improve physicians’ ability to predict cardiac events. This work automatically detects soft plaques in CTA imagery using active contours driven by spatially localized probabilistic models. Plaques are identified by simultaneously segmenting the vessel from the inside-out and the outside-in using carefully chosen localized energies [[Projects:SoftPlaqueDetection|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> Soft Plaque Detection and Automatic Vessel Segmentation. PMMIA Workshop in MICCAI, Sep. 2009. | ||
| + | |- | ||
| + | |||
| + | | | [[Image:3D_Segmentation_LA.png|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:SegmentationEndocardialWall|Segmentation of the Left Atrial Wall for Atrial Fibrillation Ablation Therapy]] == | ||
| + | |||
| + | Magnetic resonance imaging (MRI) has been used for both pre- and and post-ablation assessment of the atrial wall. MRI can aid in selecting the right candidate for the ablation procedure and assessing post-ablation scar formations. Image processing techniques can be used for automatic segmentation of the atrial wall, which facilitates an accurate statistical assessment of the region. As a first step towards the general solution to the computer-assisted segmentation of the left atrial wall, in this research we propose a shape-based image segmentation framework to segment the endocardial wall of the left atrium.[[Projects:SegmentationEpicardialWall|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> Y. Gao, B. Gholami, R. S. MacLeod, J, Blauer, W. M. Haddad, and A. R. Tannenbaum, Segmentation of the Endocardial Wall of the Left Atrium using Local Region-Based Active Contours and Statistical Shape Learning, SPIE Medical Imaging, San Diego, CA, 2010. | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:Pain1.JPG|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:PainAssessment|Agitation and Pain Assessment Using Digital Imaging]] == | ||
| + | |||
| + | Pain assessment in patients who are unable to verbally | ||
| + | communicate with medical staff is a challenging problem | ||
| + | in patient critical care. The fundamental limitations in sedation | ||
| + | and pain assessment in the intensive care unit (ICU) stem | ||
| + | from subjective assessment criteria, rather than quantifiable, | ||
| + | measurable data for ICU sedation and analgesia. This often | ||
| + | results in poor quality and inconsistent treatment of patient | ||
| + | agitation and pain from nurse to nurse. Recent advancements in | ||
| + | pattern recognition techniques using a relevance vector machine | ||
| + | algorithm can assist medical staff in assessing sedation and pain | ||
| + | by constantly monitoring the patient and providing the clinician | ||
| + | with quantifiable data for ICU sedation. In this paper, we show | ||
| + | that the pain intensity assessment given by a computer classifier | ||
| + | has a strong correlation with the pain intensity assessed by | ||
| + | expert and non-expert human examiners.[[Projects:PainAssessment|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> B. Gholami, W. M. Haddad, and A. Tannenbaum, “Relevance Vector Machine Learning for Neonate Pain Intensity Assessment Using Digital Imaging,” IEEE Trans. Biomed. Eng., vol. 57, pp. 1457-1466. | ||
| + | |||
| + | B. Gholami, W. M. Haddad, and A. R. Tannenbaum, “Agitation and Pain Assessment Using Digital Imaging,” Proc. IEEE Eng. Med. Biolog. Conf., Minneapolis, MN, pp. 2176-2179, 2009 (Awarded National Institute of Biomedical Imaging and Bioengineering/National Institute of Health Student Travel Fellowship). | ||
| + | |||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:MultiObjSeg.png|200px|]] | ||
| + | | | | ||
| + | |||
| + | == [[RobustStatisticsSegmentation|Simultaneous Multiple Object Segmentation using Robust Statistics Features ]] == | ||
| + | |||
| + | Multiple objects are segmented simultaneously using several interactive active contours based on the feature image which utilizes the robust statistics of the image. [[RobustStatisticsSegmentation|More...]] | ||
| + | |||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:ShapeBasePstSegSlicer.png|200px|]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:ProstateSegmentation|Prostate Segmentation]] == | ||
| + | |||
| + | The 3D prostate MRI images are collected by collaborators at Queen’s University. With a little manual initialization, the algorithm provided the results give to the left. The method mainly uses Random Walk algorithm. [[Projects:ProstateSegmentation|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> Y. Gao, A. Tannenbaum; Shape based MRI prostate image segmentation using local information driven directional distance Bayesian method. SPIE Medical Imaging 2010. | ||
| + | |||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:ProstateRegSupineToProneInParaview.png|200px|]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:pfPtSetImgReg|Particle Filter Registration of Medical Imagery]] == | ||
| + | |||
| + | 3D volumetric image registration is performed. The method is based on registering the images through point sets, which is able to handle long distance between as well as registration between Supine and Prone pose prostate. [[Projects:pfPtSetImgReg|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> Y. Gao, R. Sandhu, G. Fichtinger, A. Tannenbaum; A coupled global registration and segmentation framework with application to magnetic resonance prostate imagery. IEEE TMI vol.29, pp1781, 2010 | ||
| + | . | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:GT-DWI-Reorientation-1.jpg|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:DWIReorientation|Re-Orientation Approach for Segmentation of DW-MRI]] == | ||
| + | |||
| + | This work proposes a methodology to segment tubular fiber bundles from diffusion weighted magnetic resonance images (DW-MRI). Segmentation is simplified by locally reorienting diffusion information based on large-scale fiber bundle geometry. [[Projects:DWIReorientation|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> Near-tubular fiber bundle segmentation for diffusion weighted imaging: segmentation through frame reorientation. Neuroimage, Mar 2009. | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:GTTubSurfaceSeg-Img1.png|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:TubularSurfaceSegmentation|Tubular Surface Segmentation Framework]] == | ||
| + | |||
| + | We have proposed a new model for tubular surfaces that transforms the problem of detecting a surface in 3D space, to detecting a curve in 4D space. Besides allowing us to impose a "soft" tubular shape prior, this also leads to computational efficiency over conventional surface segmentation approaches. [[Projects:TubularSurfaceSegmentation|More...]] | ||
| + | |||
| + | |||
| + | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi, M. Kubicki and A. Tannenbaum. Tubular Surface Evolution for Segmentation of the Cingulum Bundle From DW-MRI. September 2008. Proceedings of the Second Workshop on Mathematical Foundations of Computational Anatomy (MFCA'08), Int Conf Med Image Comput Comput Assist Interv. 2008. | ||
| + | |||
| + | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi, A. Stillman and A. Tannenbaum. Vessel Segmentation with Automatic Centerline Extraction using Tubular Surface Segmentation. September 2009. Proceedings of the Workshop on Cardiac Interventional Imaging and Biophysical Modelling (CI2BM'09), Int Conf Med Image Comput Comput Assist Interv. 2009. | ||
| + | |||
| + | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi and A. Tannenbaum. Tubular Surface Segmentation for Extracting Anatomical Structures from Medical Imagery (in submission). IEEE Transactions on Medical Imaging. | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:GT-PopStudyVis OnCBs Case19-View2.jpg|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:TubularSurfaceSegmentationPopStudy|Group Study on DW-MRI using the Tubular Surface Model]] == | ||
| + | |||
| + | We have proposed a new framework for performing group studies on DW-MRI data sets using the Tubular Surface Model of Mohan et al. We successfully apply this framework to discriminating schizophrenic cases from normal controls, as well as towards visualizing the regions of the Cingulum Bundle that are affected by Schizophrenia. [[Projects:TubularSurfaceSegmentationPopStudy|More...]] | ||
| + | |||
| + | |||
| + | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi, M. Kubicki, D. Terry and A. Tannenbaum. Population Analysis of the Cingulum Bundle using the Tubular Surface Model for Schizophrenia Detection. SPIE Medical Imaging 2010. | ||
| + | |||
| + | <font color="red">'''New: '''</font> V. Mohan, G. Sundaramoorthi, M. Kubicki and A. Tannenbaum. Population Analysis of neural fiber bundles towards schizophrenia detection and characterization, using the Tubular Surface model. Neuroimage (in preparation) | ||
| + | |||
| + | |||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:Model3D_upTrans.png|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:MGH-HeadAndNeck-RT|Adaptive Radiotherapy for head, neck and thorax]] == | ||
| + | |||
| + | We proposed an algorithm to include prior knowledge in previously segmented anatomical structures to help in the segmentation of the next structure. This will add enough prior information to allow the Graph Cuts algorithm to segment structures with fuzzy boundaries. [[Projects:MGH-HeadAndNeck-RT|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> I. Kolesov, V. Mohan, G. Sharp and A. Tannenbaum. Coupled Segmentation for Anatomical Structures by Combining Shape and Relational Spatial Information. MTNS 2010. | ||
| + | |||
| − | |||
|- | |- | ||
| + | | | [[Image:Circle seg.PNG|200px|]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:KPCASegmentation|Kernel PCA for Segmentation]] == | ||
| + | |||
| + | Segmentation performances using active contours can be drastically improved if the possible shapes of the object of interest are learnt. The goal of this work is to use Kernel PCA to learn shape priors. Kernel PCA allows for learning non linear dependencies in data sets, leading to more robust shape priors. [[Projects:KPCASegmentation|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> S. Dambreville, Y. Rathi, and A. Tannenbaum. A Framework for Image Segmentation using Image Shape Models and Kernel PCA Shape Priors. IEEE Trans Pattern Anal Mach Intell. 2008 Aug;30(8):1385-99 | ||
| + | |||
| + | |- | ||
| | [[Image:ZoomedResultWithModel.png|200px]] | | | [[Image:ZoomedResultWithModel.png|200px]] | ||
| | | | | | ||
| Line 30: | Line 180: | ||
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:P1_small.png|200px|]] |
| + | | | | ||
| + | |||
| + | == [[Projects:LabelSpace|Label Space: A Coupled Multi-Shape Representation]] == | ||
| + | |||
| + | Many techniques for multi-shape representation may often develop inaccuracies stemming from either approximations or inherent variation. Label space is an implicit representation that offers unbiased algebraic manipulation and natural expression of label uncertainty. We demonstrate smoothing and registration on multi-label brain MRI. [[Projects:LabelSpace|More...]] | ||
| + | |||
| + | <font color="red">'''New: '''</font> J. Malcolm, Y. Rathi, A. Tannenbaum. "Label Space: A Multi-Object Shape Representation." In Combinatorial Image Analysis, 2008. | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:BasePair3DModel.JPG|200px|]] | ||
| | | | | | ||
| − | == [[Projects: | + | == [[Projects:NonParametricClustering|Non Parametric Clustering for Biomolecular Structural Analysis]] == |
| − | + | High accuracy imaging and image processing techniques allow for collecting structural information of biomolecules with atomistic accuracy. Direct interpretation of the dynamics and the functionality of these structures with physical models, is yet to be developed. Clustering of molecular conformations into classes seems to be the first stage in recovering the formation and the functionality of these molecules. [[Projects:NonParametricClustering|More...]] | |
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> E. Hershkovits, A. Tannenbaum, and R. Tannenbaum. Adsorption of Block Copolymers from Selective Solvents on Curved Surfaces. Macromolecules. 2008. |
|- | |- | ||
| Line 50: | Line 211: | ||
<font color="red">'''New: '''</font> R. Sandhu, S. Dambreville, A. Tannenbaum. Particle Filtering for Registration of 2D and 3D Point Sets with Stochastic Dynamics. In CVPR, 2008. | <font color="red">'''New: '''</font> R. Sandhu, S. Dambreville, A. Tannenbaum. Particle Filtering for Registration of 2D and 3D Point Sets with Stochastic Dynamics. In CVPR, 2008. | ||
| + | |||
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:Results brain sag.JPG|200px]] |
| | | | | | ||
| − | == [[Projects: | + | == [[Projects:OptimalMassTransportRegistration|Optimal Mass Transport Registration]] == |
| + | |||
| + | The goal of this project is to implement a computationaly efficient Elastic/Non-rigid Registration algorithm based on the Monge-Kantorovich theory of optimal mass transport for 3D Medical Imagery. Our technique is based on Multigrid and Multiresolution techniques. This method is particularly useful because it is parameter free and utilizes all of the grayscale data in the image pairs in a symmetric fashion and no landmarks need to be specified for correspondence. [[Projects:OptimalMassTransportRegistration|More...]] | ||
| − | + | <font color="red">'''New: '''</font> Eldad Haber, Tauseef Rehman, and Allen Tannenbaum. An Efficient Numerical Method for the Solution of the L2 Optimal Mass Transfer Problem. In submission - SIAM Journal of Scientific Computing, 2009. | |
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> Tauseef Rehman, Eldad Haber, Gallagher Pryor, and Allen Tannenbaum. Fast Optimal Mass Transport for 2D Image Registration and Morphing. Accepted for - Elsevier Journal of Image and Vision Computing, 2009. |
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:Gatech caudateBands.PNG|200px]] |
| | | | | | ||
| − | == [[Projects: | + | == [[Projects:MultiscaleShapeSegmentation|Multiscale Shape Segmentation Techniques]] == |
| − | + | To represent multiscale variations in a shape population in order to drive the segmentation of deep brain structures, such as the caudate nucleus or the hippocampus. [[Projects:MultiscaleShapeSegmentation|More...]] | |
| − | |||
| − | |||
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:Caudate Nucleus Denoising.JPG|200px|]] |
| | | | | | ||
| − | == [[Projects: | + | == [[Projects:WaveletShrinkage|Wavelet Shrinkage for Shape Analysis]] == |
| − | + | Shape analysis has become a topic of interest in medical imaging since local variations of a shape could carry relevant information about a disease that may affect only a portion of an organ. We developed a novel wavelet-based denoising and compression statistical model for 3D shapes. [[Projects:WaveletShrinkage|More...]] | |
| − | |||
| − | |||
|- | |- | ||
| Line 93: | Line 253: | ||
We present a novel method of statistical surface-based morphometry based on the use of non-parametric permutation tests and a spherical wavelet (SWC) shape representation. [[Projects:MultiscaleShapeAnalysis|More...]] | We present a novel method of statistical surface-based morphometry based on the use of non-parametric permutation tests and a spherical wavelet (SWC) shape representation. [[Projects:MultiscaleShapeAnalysis|More...]] | ||
| − | + | |- | |
| + | |||
| + | | | [[Image:Dlpfc1.jpg|200px|]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:RuleBasedDLPFCSegmentation|Rule-Based DLPFC Segmentation]] == | ||
| + | |||
| + | In this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the dorsolateral prefrontal cortex. [[Projects:RuleBasedDLPFCSegmentation|More...]] | ||
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:Striatum1.png|200px|]] |
| | | | | | ||
| − | == [[Projects: | + | == [[Projects:RuleBasedStriatumSegmentation|Rule-Based Striatum Segmentation]] == |
| + | |||
| + | In this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the striatum. [[Projects:RuleBasedStriatumSegmentation|More...]] | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:Brain-flat.PNG|200px]] | ||
| + | | | | ||
| − | + | == [[Projects:ConformalFlatteningRegistration|Conformal Flattening (inactive)]] == | |
| − | + | The goal of this project is for better visualizing and computation of neural activity from fMRI brain imagery. Also, with this technique, shapes can be mapped to shperes for shape analysis, registration or other purposes. Our technique is based on conformal mappings which map genus-zero surface: in fmri case cortical or other surfaces, onto a sphere in an angle preserving manner. [[Projects:ConformalFlatteningRegistration|More...]] | |
|- | |- | ||
| Line 115: | Line 289: | ||
The goal of this work is to develop blood vessel segmentation techniques for 3D MRI and CT data. The methods have been applied to coronary arteries and portal veins, with promising results. [[Projects:BloodVesselSegmentation|More...]] | The goal of this work is to develop blood vessel segmentation techniques for 3D MRI and CT data. The methods have been applied to coronary arteries and portal veins, with promising results. [[Projects:BloodVesselSegmentation|More...]] | ||
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> V.Mohan, G. Sundaramoorthi, A. Stillman and A. Tannenbaum. Vessel Segmentation with Automatic Centerline Extraction Using Tubular Tree Segmentation. CI2BM at MICCAI 2009, September 2009. |
|- | |- | ||
| Line 125: | Line 299: | ||
This ITK filter is a segmentation algorithm that utilizes Bayes's Rule along with an affine-invariant anisotropic smoothing filter. [[Projects:KnowledgeBasedBayesianSegmentation|More...]] | This ITK filter is a segmentation algorithm that utilizes Bayes's Rule along with an affine-invariant anisotropic smoothing filter. [[Projects:KnowledgeBasedBayesianSegmentation|More...]] | ||
| − | |||
| − | |||
|- | |- | ||
| Line 137: | Line 309: | ||
New stochastic methods for implementing curvature driven flows for various medical tasks such as segmentation. [[Projects:StochasticMethodsSegmentation|More...]] | New stochastic methods for implementing curvature driven flows for various medical tasks such as segmentation. [[Projects:StochasticMethodsSegmentation|More...]] | ||
| − | + | |- | |
| + | |||
| + | | | [[Image:GT-SulciOutlining1.jpg|200px]] | ||
| + | | | | ||
| + | |||
| + | == [[Projects:SulciOutlining|Automatic Outlining of sulci on the brain surface]] == | ||
| + | |||
| + | We present a method to automatically extract certain key features on a surface. We apply this technique to outline sulci on the cortical surface of a brain. [[Projects:SulciOutlining|More...]] | ||
|- | |- | ||
| Line 147: | Line 326: | ||
The goal of this work is to study and compare shape learning techniques. The techniques considered are Linear Principal Components Analysis (PCA), Kernel PCA, Locally Linear Embedding (LLE) and Kernel LLE. [[Projects:KPCA_LLE_KLLE_ShapeAnalysis|More...]] | The goal of this work is to study and compare shape learning techniques. The techniques considered are Linear Principal Components Analysis (PCA), Kernel PCA, Locally Linear Embedding (LLE) and Kernel LLE. [[Projects:KPCA_LLE_KLLE_ShapeAnalysis|More...]] | ||
| − | |||
| − | |||
|- | |- | ||
| Line 159: | Line 336: | ||
This Fast Marching based flow was added to Slicer 2. [[Projects:StatisticalSegmentationSlicer2|More...]] | This Fast Marching based flow was added to Slicer 2. [[Projects:StatisticalSegmentationSlicer2|More...]] | ||
| − | | | + | |} |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 20:36, 16 October 2011
Home < Algorithm:GATechBack to NA-MIC Algorithms
Overview of Boston University Algorithms (PI: Allen Tannenbaum)
At Boston University, we are broadly interested in a range of mathematical image analysis algorithms for segmentation, registration, diffusion-weighted MRI analysis, and statistical analysis. For many applications, we cast the problem in an energy minimization framework wherein we define a partial differential equation whose numeric solution corresponds to the desired algorithmic outcome. The following are many examples of PDE techniques applied to medical image analysis.
Boston University Projects

|
Multi-scale Shape Representation and Segmentation With Applications to RadiotherapyWe present in this work a multiscale representation for shapes with arbitrary topology, and a method to segment the target organ/tissue from medical images having very low contrast with respect to surrounding regions using multiscale shape information and local image features. In a number of previous papers, shape knowledge was incorporated by first constructing a shape space from training data, and then constraining the segmentation process to be within the resulting shape space. However, such an approach has certain limitations including the fact that small scale shape variances may be overwhelmed by those on larger scale, and therefore the local shape information is lost. In this work, first we handle this problem by providing a multiscale shape representation using the wavelet transform. Consequently, the shape variances captured by the statistical learning step are also represented at various scales. In doing so, not only is the diversity of shape enriched, but also small scale changes are nicely captured. More... |
|
Soft Plaque Detection in CTA ImageryThe ability to detect and measure non-calcified plaques (also known as soft plaques) may improve physicians’ ability to predict cardiac events. This work automatically detects soft plaques in CTA imagery using active contours driven by spatially localized probabilistic models. Plaques are identified by simultaneously segmenting the vessel from the inside-out and the outside-in using carefully chosen localized energies More... New: Soft Plaque Detection and Automatic Vessel Segmentation. PMMIA Workshop in MICCAI, Sep. 2009. |
|
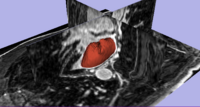
Segmentation of the Left Atrial Wall for Atrial Fibrillation Ablation TherapyMagnetic resonance imaging (MRI) has been used for both pre- and and post-ablation assessment of the atrial wall. MRI can aid in selecting the right candidate for the ablation procedure and assessing post-ablation scar formations. Image processing techniques can be used for automatic segmentation of the atrial wall, which facilitates an accurate statistical assessment of the region. As a first step towards the general solution to the computer-assisted segmentation of the left atrial wall, in this research we propose a shape-based image segmentation framework to segment the endocardial wall of the left atrium.More... New: Y. Gao, B. Gholami, R. S. MacLeod, J, Blauer, W. M. Haddad, and A. R. Tannenbaum, Segmentation of the Endocardial Wall of the Left Atrium using Local Region-Based Active Contours and Statistical Shape Learning, SPIE Medical Imaging, San Diego, CA, 2010. |
|
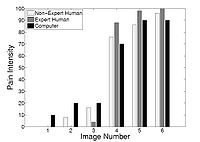
Agitation and Pain Assessment Using Digital ImagingPain assessment in patients who are unable to verbally communicate with medical staff is a challenging problem in patient critical care. The fundamental limitations in sedation and pain assessment in the intensive care unit (ICU) stem from subjective assessment criteria, rather than quantifiable, measurable data for ICU sedation and analgesia. This often results in poor quality and inconsistent treatment of patient agitation and pain from nurse to nurse. Recent advancements in pattern recognition techniques using a relevance vector machine algorithm can assist medical staff in assessing sedation and pain by constantly monitoring the patient and providing the clinician with quantifiable data for ICU sedation. In this paper, we show that the pain intensity assessment given by a computer classifier has a strong correlation with the pain intensity assessed by expert and non-expert human examiners.More... New: B. Gholami, W. M. Haddad, and A. Tannenbaum, “Relevance Vector Machine Learning for Neonate Pain Intensity Assessment Using Digital Imaging,” IEEE Trans. Biomed. Eng., vol. 57, pp. 1457-1466. B. Gholami, W. M. Haddad, and A. R. Tannenbaum, “Agitation and Pain Assessment Using Digital Imaging,” Proc. IEEE Eng. Med. Biolog. Conf., Minneapolis, MN, pp. 2176-2179, 2009 (Awarded National Institute of Biomedical Imaging and Bioengineering/National Institute of Health Student Travel Fellowship).
|

|
Simultaneous Multiple Object Segmentation using Robust Statistics FeaturesMultiple objects are segmented simultaneously using several interactive active contours based on the feature image which utilizes the robust statistics of the image. More...
|
|
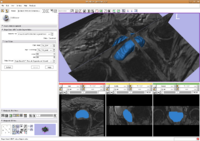
Prostate SegmentationThe 3D prostate MRI images are collected by collaborators at Queen’s University. With a little manual initialization, the algorithm provided the results give to the left. The method mainly uses Random Walk algorithm. More... New: Y. Gao, A. Tannenbaum; Shape based MRI prostate image segmentation using local information driven directional distance Bayesian method. SPIE Medical Imaging 2010.
|

|
Particle Filter Registration of Medical Imagery3D volumetric image registration is performed. The method is based on registering the images through point sets, which is able to handle long distance between as well as registration between Supine and Prone pose prostate. More... New: Y. Gao, R. Sandhu, G. Fichtinger, A. Tannenbaum; A coupled global registration and segmentation framework with application to magnetic resonance prostate imagery. IEEE TMI vol.29, pp1781, 2010 . |

|
Re-Orientation Approach for Segmentation of DW-MRIThis work proposes a methodology to segment tubular fiber bundles from diffusion weighted magnetic resonance images (DW-MRI). Segmentation is simplified by locally reorienting diffusion information based on large-scale fiber bundle geometry. More... New: Near-tubular fiber bundle segmentation for diffusion weighted imaging: segmentation through frame reorientation. Neuroimage, Mar 2009. |

|
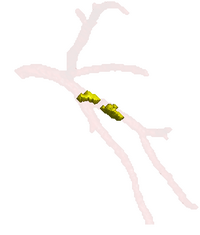
Tubular Surface Segmentation FrameworkWe have proposed a new model for tubular surfaces that transforms the problem of detecting a surface in 3D space, to detecting a curve in 4D space. Besides allowing us to impose a "soft" tubular shape prior, this also leads to computational efficiency over conventional surface segmentation approaches. More...
New: V. Mohan, G. Sundaramoorthi, A. Stillman and A. Tannenbaum. Vessel Segmentation with Automatic Centerline Extraction using Tubular Surface Segmentation. September 2009. Proceedings of the Workshop on Cardiac Interventional Imaging and Biophysical Modelling (CI2BM'09), Int Conf Med Image Comput Comput Assist Interv. 2009. New: V. Mohan, G. Sundaramoorthi and A. Tannenbaum. Tubular Surface Segmentation for Extracting Anatomical Structures from Medical Imagery (in submission). IEEE Transactions on Medical Imaging. |
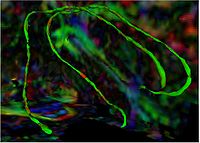
|
Group Study on DW-MRI using the Tubular Surface ModelWe have proposed a new framework for performing group studies on DW-MRI data sets using the Tubular Surface Model of Mohan et al. We successfully apply this framework to discriminating schizophrenic cases from normal controls, as well as towards visualizing the regions of the Cingulum Bundle that are affected by Schizophrenia. More...
New: V. Mohan, G. Sundaramoorthi, M. Kubicki and A. Tannenbaum. Population Analysis of neural fiber bundles towards schizophrenia detection and characterization, using the Tubular Surface model. Neuroimage (in preparation)
|
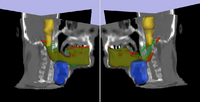
|
Adaptive Radiotherapy for head, neck and thoraxWe proposed an algorithm to include prior knowledge in previously segmented anatomical structures to help in the segmentation of the next structure. This will add enough prior information to allow the Graph Cuts algorithm to segment structures with fuzzy boundaries. More... New: I. Kolesov, V. Mohan, G. Sharp and A. Tannenbaum. Coupled Segmentation for Anatomical Structures by Combining Shape and Relational Spatial Information. MTNS 2010.
|
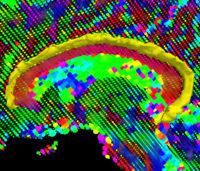
Kernel PCA for SegmentationSegmentation performances using active contours can be drastically improved if the possible shapes of the object of interest are learnt. The goal of this work is to use Kernel PCA to learn shape priors. Kernel PCA allows for learning non linear dependencies in data sets, leading to more robust shape priors. More... New: S. Dambreville, Y. Rathi, and A. Tannenbaum. A Framework for Image Segmentation using Image Shape Models and Kernel PCA Shape Priors. IEEE Trans Pattern Anal Mach Intell. 2008 Aug;30(8):1385-99 | |
|
Geodesic Tractography SegmentationIn this work, we provide an energy minimization framework which allows one to find fiber tracts and volumetric fiber bundles in brain diffusion-weighted MRI (DW-MRI). More... New: J. Melonakos, E. Pichon, S. Angenet, and A. Tannenbaum. Finsler Active Contours. IEEE Transactions on Pattern Analysis and Machine Intelligence, March 2008, Vol 30, Num 3. |

|
Label Space: A Coupled Multi-Shape RepresentationMany techniques for multi-shape representation may often develop inaccuracies stemming from either approximations or inherent variation. Label space is an implicit representation that offers unbiased algebraic manipulation and natural expression of label uncertainty. We demonstrate smoothing and registration on multi-label brain MRI. More... New: J. Malcolm, Y. Rathi, A. Tannenbaum. "Label Space: A Multi-Object Shape Representation." In Combinatorial Image Analysis, 2008. |
|
Non Parametric Clustering for Biomolecular Structural AnalysisHigh accuracy imaging and image processing techniques allow for collecting structural information of biomolecules with atomistic accuracy. Direct interpretation of the dynamics and the functionality of these structures with physical models, is yet to be developed. Clustering of molecular conformations into classes seems to be the first stage in recovering the formation and the functionality of these molecules. More... New: E. Hershkovits, A. Tannenbaum, and R. Tannenbaum. Adsorption of Block Copolymers from Selective Solvents on Curved Surfaces. Macromolecules. 2008. |
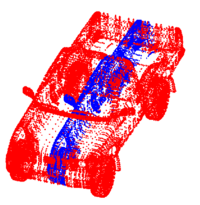
|
Point Set Rigid RegistrationIn this work, we propose a particle filtering approach for the problem of registering two point sets that differ by a rigid body transformation. Experimental results are provided that demonstrate the robustness of the algorithm to initialization, noise, missing structures or differing point densities in each sets, on challenging 2D and 3D registration tasks. More... New: R. Sandhu, S. Dambreville, A. Tannenbaum. Particle Filtering for Registration of 2D and 3D Point Sets with Stochastic Dynamics. In CVPR, 2008.
|

|
Optimal Mass Transport RegistrationThe goal of this project is to implement a computationaly efficient Elastic/Non-rigid Registration algorithm based on the Monge-Kantorovich theory of optimal mass transport for 3D Medical Imagery. Our technique is based on Multigrid and Multiresolution techniques. This method is particularly useful because it is parameter free and utilizes all of the grayscale data in the image pairs in a symmetric fashion and no landmarks need to be specified for correspondence. More... New: Eldad Haber, Tauseef Rehman, and Allen Tannenbaum. An Efficient Numerical Method for the Solution of the L2 Optimal Mass Transfer Problem. In submission - SIAM Journal of Scientific Computing, 2009. New: Tauseef Rehman, Eldad Haber, Gallagher Pryor, and Allen Tannenbaum. Fast Optimal Mass Transport for 2D Image Registration and Morphing. Accepted for - Elsevier Journal of Image and Vision Computing, 2009. |
|
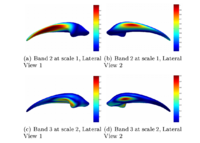
Multiscale Shape Segmentation TechniquesTo represent multiscale variations in a shape population in order to drive the segmentation of deep brain structures, such as the caudate nucleus or the hippocampus. More... |

|
Wavelet Shrinkage for Shape AnalysisShape analysis has become a topic of interest in medical imaging since local variations of a shape could carry relevant information about a disease that may affect only a portion of an organ. We developed a novel wavelet-based denoising and compression statistical model for 3D shapes. More... |
|
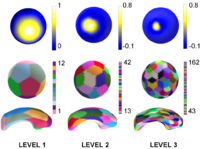
Multiscale Shape AnalysisWe present a novel method of statistical surface-based morphometry based on the use of non-parametric permutation tests and a spherical wavelet (SWC) shape representation. More... |
|
Rule-Based DLPFC SegmentationIn this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the dorsolateral prefrontal cortex. More... |
|
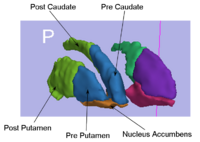
Rule-Based Striatum SegmentationIn this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the striatum. More... |

|
Conformal Flattening (inactive)The goal of this project is for better visualizing and computation of neural activity from fMRI brain imagery. Also, with this technique, shapes can be mapped to shperes for shape analysis, registration or other purposes. Our technique is based on conformal mappings which map genus-zero surface: in fmri case cortical or other surfaces, onto a sphere in an angle preserving manner. More... |
|
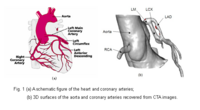
Blood Vessel SegmentationThe goal of this work is to develop blood vessel segmentation techniques for 3D MRI and CT data. The methods have been applied to coronary arteries and portal veins, with promising results. More... New: V.Mohan, G. Sundaramoorthi, A. Stillman and A. Tannenbaum. Vessel Segmentation with Automatic Centerline Extraction Using Tubular Tree Segmentation. CI2BM at MICCAI 2009, September 2009. |
|
Knowledge-Based Bayesian SegmentationThis ITK filter is a segmentation algorithm that utilizes Bayes's Rule along with an affine-invariant anisotropic smoothing filter. More... |
|
Stochastic Methods for SegmentationNew stochastic methods for implementing curvature driven flows for various medical tasks such as segmentation. More... |
|
Automatic Outlining of sulci on the brain surfaceWe present a method to automatically extract certain key features on a surface. We apply this technique to outline sulci on the cortical surface of a brain. More... |
KPCA, LLE, KLLE Shape AnalysisThe goal of this work is to study and compare shape learning techniques. The techniques considered are Linear Principal Components Analysis (PCA), Kernel PCA, Locally Linear Embedding (LLE) and Kernel LLE. More... | |

|
Statistical/PDE Methods using Fast Marching for SegmentationThis Fast Marching based flow was added to Slicer 2. More... |