|
|
| (15 intermediate revisions by one other user not shown) |
| Line 1: |
Line 1: |
| | Back to [[NA-MIC_Collaborations:New|NA-MIC Collaborations]] | | Back to [[NA-MIC_Collaborations:New|NA-MIC Collaborations]] |
| | __NOTOC__ | | __NOTOC__ |
| − | == Diffusion Image Analysis == | + | = NA-MIC Internal Collaborations = |
| | | | |
| − | === Fiber Tract Extraction and Analysis === | + | {| cellpadding="10" border="1" |
| | + | | style="width:33%" | [[Image:CingulumAllSubjectsFibers.png|300px]] |
| | + | | style="width:33%" | [[Image:Sulcaldepth.png|300px]] |
| | + | | style="width:33%" | [[Image:Mit_fmri_clustering_parcellation2_xsub.png|300px]] |
| | | | |
| − | {| cellpadding="10"
| + | |-align="center" |
| − | | style="width:15%" | [[Image:ZoomedResultWithModel.png|200px]]
| |
| − | | style="width:85%" |
| |
| − | | |
| − | == [[Projects:GeodesicTractographySegmentation|Geodesic Tractography Segmentation]] ==
| |
| − | | |
| − | In this work, we provide an energy minimization framework which allows one to find fiber tracts and volumetric fiber bundles in brain diffusion-weighted MRI (DW-MRI). [[Projects:GeodesicTractographySegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> J. Melonakos, E. Pichon, S. Angenet, and A. Tannenbaum. Finsler Active Contours. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2007.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:MIT_DTI_JointSegReg_atlas3D.jpg|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:DTIFiberRegistration|Joint Registration and Segmentation of DWI Fiber Tractography]] ==
| |
| − | | |
| − | The goal of this work is to jointly register and cluster DWI fiber tracts obtained from a group of subjects. [[Projects:DTIFiberRegistration|More...]]
| |
| − | | |
| − | <font color="red">'''New:'''</font> U. Ziyan, M. R. Sabuncu, W. E. L. Grimson, Carl-Fredrik Westin. A Robust Algorithm for Fiber-Bundle Atlas Construction. MMBIA 2007.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:Thalamus_algo_outline.png|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:DTISegmentation|DTI-based Segmentation]] ==
| |
| − | | |
| − | Unlike conventional MRI, DTI provides adequate contrast to segment the thalamic nuclei, which are gray matter structures. [[Projects:DTISegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New:'''</font> Ulas Ziyan, David Tuch, Carl-Fredrik Westin. Segmentation of Thalamic Nuclei from DTI using Spectral Clustering. Accepted to MICCAI 2006.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:FiberTracts-angle.jpg|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:DTIVolumetricWhiteMatterConnectivity|DTI Volumetric White Matter Connectivity]] ==
| |
| − | | |
| − | We have developed a PDE-based approach to white matter connectivity from DTI that is founded on the principal of minimal paths through the tensor volume. Our method computes a volumetric representation of a white matter tract given two endpoint regions. We have also developed statistical methods for quantifying the full tensor data along these pathways, which should be useful in clinical studies using DT-MRI. [[Projects:DTIVolumetricWhiteMatterConnectivity|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> PT Fletcher, R Tao, W-K Jeong, RT Whitaker, A volumetric approach to quantifying region-to-region white matter connectivity in diffusion tensor MRI, IPMI 2007, pp. 346-358.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:ConnectivityMap.png|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:DTIStochasticTractography|Stochastic Tractography]] ==
| |
| − | | |
| − | This work calculates posterior distributions of white matter fiber tract parameters given diffusion observations in a DWI volume. [[Projects:DTIStochasticTractography|More...]]
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:CingulumAllSubjectsFibers.png|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:DTIClustering|DTI Fiber Clustering and Fiber-Based Analysis]] ==
| |
| − | | |
| − | The goal of this project is to provide structural description of the white matter architecture as a partition into coherent fiber bundles and clusters, and to use these bundles for quantitative measurement. [[Projects:DTIClustering|More...]]
| |
| − | | |
| − | <font color="red">'''New:'''</font> Monica E. Lemmond, Lauren J. O'Donnell, Stephen Whalen, Alexandra J. Golby. Characterizing Diffusion Along White Matter Tracts Affected by Primary Brain Tumors. Accepted to HBM 2007.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:Models.jpg|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:DTIModeling|Fiber Tract Modeling, Clustering, and Quantitative Analysis]] ==
| |
| − | | |
| − | The goal of this work is to model the shape of the fiber bundles and use this model discription in clustering and statistical analysis of fiber tracts. [[Projects:DTIModeling|More...]]
| |
| − | | |
| − | <font color="red">'''New:'''</font>
| |
| − | M. Maddah, W. E. L. Grimson, S. K. Warfield, W. M. Wells, A Unified Framework for Clustering and Quantitative Analysis of White Matter Fiber Tracts. Medical Image Analysis, in press.
| |
| − | | |
| − | M. Maddah, W. M. Wells, S. K. Warfield, C.-F. Westin, and W. E. L. Grimson, Probabilistic Clustering and Quantitative Analysis of White Matter Fiber Tracts, IPMI 2007, Netherlands.
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Fractional Anisotropy Analysis ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" | [[Image:DTIQuantitativeAnalysis.png|200px]]
| |
| − | | style="width:85%" |
| |
| − | | |
| − | == [[Projects:DTIQuantitativeTractAnalysis|Quantitative Analysis of Fiber Tract Bundles]] ==
| |
| − | | |
| − | DT-MRI tractography can be used as a coordinate system for computing statistics of diffusion tensor data. The quantitative analysis of diffusion tensors takes into account the space of tensor measurements using a nonlinear Riemannian symmetric space framework. Tracts of interest are represented as a medial spline attributed with cross-sectional statistics. [[Projects:DTIQuantitativeTractAnalysis|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Gilmore J, Lin W, Corouge I, Vetsa Y, Smith J, Kang C, Gu H, Hamer R, Lieberman J, Gerig G. Early Postnatal Development of Corpus Callosum and Corticospinal White Matter Assessed with Quantitative Tractography. AJNR Am J Neuroradiol. 2007.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:DTINoiseStatistics.png|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:DTINoiseStatistics|Influence of Imaging Noise on DTI Statistics]] ==
| |
| − | | |
| − | Clinical acquisition of diffusion weighted images with high signal to noise ratio remains a challenge. The goal of this project is to understand the impact of MR noise on quantiative statistics of diffusion properties such as anisotropy measures, trace, etc. [[Projects:DTINoiseStatistics|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Casey Goodlett, P. Thomas Fletcher, Weili Lin, Guido Gerig. Quantification of measurement error in DTI: Theoretical predictions and validation, MICCAI 2007.
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Path of Interest Analysis ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Validation ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Algorithm/Software Infrastructure ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |}
| |
| − | | |
| − | == Structural Image Analysis ==
| |
| − | | |
| − | === Image Segmentation ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" | [[Image:Fig67.png|200px|]]
| |
| − | | style="width:85%" |
| |
| − | | |
| − | == [[Projects:KnowledgeBasedBayesianSegmentation|Knowledge-Based Bayesian Segmentation]] ==
| |
| − | | |
| − | This ITK filter is a segmentation algorithm that utilizes Bayes's Rule along with an affine-invariant anisotropic smoothing filter. [[Projects:KnowledgeBasedBayesianSegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> J. Melonakos, Y. Gao, and A. Tannenbaum. Tissue Tracking: Applications for Brain MRI Classification. SPIE Medical Imaging, 2007.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:Striatum1.png|200px|]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:RuleBasedStriatumSegmentation|Rule-Based Striatum Segmentation]] ==
| |
| − | | |
| − | In this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the striatum. [[Projects:RuleBasedStriatumSegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Al-Hakim, et al. Parcellation of the Striatum. SPIE MI 2007.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:Dlpfc1.jpg|200px|]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:RuleBasedDLPFCSegmentation|Rule-Based DLPFC Segmentation]] ==
| |
| − | | |
| − | In this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the dorsolateral prefrontal cortex. [[Projects:RuleBasedDLPFCSegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Al-Hakim, et al. A Dorsolateral Prefrontal Cortex Semi-Automatic Segmenter. SPIE MI 2006.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:Gatech caudateBands.PNG|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:MultiscaleShapeSegmentation|Multiscale Shape Segmentation Techniques]] ==
| |
| − | | |
| − | To represent multiscale variations in a shape population in order to drive the segmentation of deep brain structures, such as the caudate nucleus or the hippocampus. [[Projects:MultiscaleShapeSegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Delphine Nain won the best student paper at [[MICCAI_2006|MICCAI 2006]] in the category "Segmentation and Registration" for her paper entitled "Shape-driven surface segmentation using spherical wavelets" by D. Nain, S. Haker, A. Bobick, A. Tannenbaum.
| |
| | | | |
| − | |-
| |
| − |
| |
| − | | | [[Image:Stochastic-snake.png|200px|]]
| |
| | | | | | | | |
| | + | == [[NA-MIC_Internal_Collaborations:DiffusionImageAnalysis|Diffusion Image Analysis]] == |
| | | | |
| − | == [[Projects:StochasticMethodsSegmentation|Stochastic Methods for Segmentation]] ==
| |
| − |
| |
| − | New stochastic methods for implementing curvature driven flows for various medical tasks such as segmentation. [[Projects:StochasticMethodsSegmentation|More...]]
| |
| − |
| |
| − | <font color="red">'''New: '''</font> Currently under investigation.
| |
| − |
| |
| − | |-
| |
| − |
| |
| − | | | [[Image:Gatech SlicerModel2.jpg|200px]]
| |
| | | | | | | | |
| | + | == [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|Structural Image Analysis]] == |
| | | | |
| − | == [[Projects:StatisticalSegmentationSlicer2|Statistical/PDE Methods using Fast Marching for Segmentation]] ==
| |
| − |
| |
| − | This Fast Marching based flow was added to Slicer 2. [[Projects:StatisticalSegmentationSlicer2|More...]]
| |
| − |
| |
| − | |-
| |
| − |
| |
| − | | | [[Image:brain_classify.png|200px]]
| |
| | | | | | | | |
| − | | + | == [[NA-MIC_Internal_Collaborations:fMRIAnalysis|fMRI Analysis]] == |
| − | == [[Projects:TissueClassificationWithNeighborhoodStatistics| Tissue Classification with Neighborhood Statistics]] == | |
| − | | |
| − | We have implemented an MRI tissue classification algorithm based on unsupervised non-parametric density estimation of tissue intensity classes.
| |
| − | [[Projects:TissueClassificationWithNeighborhoodStatistics|More...]]
| |
| − | | |
| − | T Tasdizen, S Awate, R Whitaker, A nonparametric, entropy-minimizing MRI tissue classification algorithm implementation using ITK, MICCAI 2005 Open-Source Workshop.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:histo_matching.jpg|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:AutomaticFullBrainSegmentation|Atlas Renormalization for Improved Brain MR Image Segmentation across Scanner Platforms]] ==
| |
| − | | |
| − | Atlas-based approaches have demonstrated the ability to automatically identify detailed brain structures from 3-D magnetic resonance (MR) brain images. Unfortunately, the accuracy of this type of method often degrades when processing data acquired on a different scanner platform or pulse sequence than the data used for the atlas training. In this paper, we improve the performance of an atlas-based whole brain segmentation method by introducing an intensity renormalization procedure that automatically adjusts the prior atlas intensity model to new input data. Validation using manually labeled test datasets has shown that the new procedure improves the segmentation accuracy (as measured by the Dice coefficient) by 10% or more for several structures including hippocampus, amygdala, caudate, and pallidum. The results verify that this new procedure reduces the sensitivity of the whole brain segmentation method to changes in scanner platforms and improves its accuracy and robustness, which can thus facilitate multicenter or multisite neuroanatomical imaging studies. [[Projects:AutomaticFullBrainSegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 4, APRIL 2007
| |
| | | | |
| | |} | | |} |
| | | | |
| − | === Image Registration === | + | = Driving Biological Problems (2007 - 2010): Roadmap Projects = |
| | | | |
| − | {| cellpadding="10" | + | {| cellpadding="10" border="1" |
| − | | style="width:15%" | [[Image:Results brain sag.JPG|200px|]] | + | | style="width:15%" | [[Image:Cingulum1.jpg|200px]] |
| | | style="width:85%" | | | | style="width:85%" | |
| | | | |
| − | == [[Projects:OptimalMassTransportRegistration|Optimal Mass Transport Registration]] == | + | == [[ProjectWeek200706:ContrastingTractographyMeasures|Contrasting Tractography Measures]] == |
| | | | |
| − | The goal of this project is to implement a computationaly efficient Elastic/Non-rigid Registration algorithm based on the Monge-Kantorovich theory of optimal mass transport for 3D Medical Imagery. Our technique is based on Multigrid and Multiresolution techniques. This method is particularly useful because it is parameter free and utilizes all of the grayscale data in the image pairs in a symmetric fashion and no landmarks need to be specified for correspondence. [[Projects:OptimalMassTransportRegistration|More...]]
| + | This project represents a new initiative to build upon a shared vision among Cores 1, 3 and 5 that the field of medical image analysis would be well served by work in the area of validation, calibration and assessment of reliability in DW-MRI image analysis. [[ProjectWeek200706:ContrastingTractographyMeasures|More...]] |
| | | | |
| − | <font color="red">'''New: '''</font> Tauseef ur Rehman, A. Tannenbaum. Multigrid Optimal Mass Transport for Image Registration and Morphing. SPIE Conference on Computational Imaging V, Jan 2007. | + | <font color="red">'''New: '''</font> [[SanteFe.Tractography.Conference|Contrasting Tractography Methods Conference]], Santa Fe, October 1-2, 2007. |
| | | | |
| | |- | | |- |
| | | | |
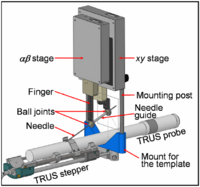
| − | | | [[Image:DTIregistration200.png|200px]] | + | | | [[Image:ProstateDiagram.png|200px]] |
| | | | | | | | |
| | | | |
| − | == [[Projects:DTIProcessingTools|Diffusion Tensor Image Processing Tools]] == | + | == [[ProjectWeek200706:BrachytherapyNeedlePositioningRobotIntegration|Brachytherapy Needle Positioning Robot Integration]] == |
| | | | |
| − | We implement the diffusion weighted image (DWI) registration model from the paper of G.K.Rohde et al. Patient head motion and eddy currents distortion cause artifacts in maps of diffusion parameters computer from DWI. This model corrects these two distortions at the same time including brightness correction.
| + | The Queen’s/Hopkins team is developing novel devices and procedures for cancer interventions, including biopsy and therapies. Our goal for the programming week is to design and start implementing software for the new MRI Brachytherapy needle positioning robot. [[ProjectWeek200706:BrachytherapyNeedlePositioningRobotIntegration|More...]] |
| | | | |
| − | <font color="red">'''New: '''</font> We have recently developed software for eddy current correction. | + | <font color="red">'''New: '''</font> Meeting at JHU on July 17-19, 2007. |
| | | | |
| | |- | | |- |
| | | | |
| − | | | [[Image:Sulcaldepth.png|200px]] | + | | | [[Image:Lupus.png|200px]] |
| | | | | | | | |
| | | | |
| − | == [[Projects:CorticalCorrespondenceWithParticleSystem|Cortical Correspondence using Particle System]] == | + | == [[DBP2:MIND:Roadmap|Brain Lesion Analysis in Neuropsychiatric Systemic Lupus Erythematosus]] == |
| | | | |
| − | In this project, we want to compute cortical correspondence on populations, using various features such as cortical structure, DTI connectivity, vascular structure, and functional data (fMRI). This presents a challenge because of the highly convoluted surface of the cortex, as well as because of the different properties of the data features we want to incorporate together. [[Projects:CorticalCorrespondenceWithParticleSystem|More...]]
| + | Our goal is to automatically, or with little or no manual human rater input, accurately tissue classify our example lupus data-set into gray, white, csf, and lesion classes. |
| | | | |
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> Participants in the Summer 2007 NA-MIC Project Week. |
| − | | |
| − | * ISBI 2007 submission
| |
| − | * Currently working with cortical data using sulcal depth and evaluating on sulcal depth/cortical thickness data.
| |
| | | | |
| | |- | | |- |
| | | | |
| − | | | [[Image:Cbg-dtiatlas-tracts.png|200px]] | + | | | [[Image:Itksnap.jpg|200px]] |
| − | | |
| |
| − | | |
| − | == [[Projects:DTIPopulationAnalysis|Population Analysis from Deformable Registration]] ==
| |
| − | | |
| − | Analysis of populations of diffusion images typically requires time-consuming manual segmentation of structures of interest to obtain correspondance for statistics. This project uses non-rigid registration of DTI images to produce a common coordinate system for hypothesis testing of diffusion properties. [[Projects:DTIPopulationAnalysis|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Command line DTI tools available as part of UNC [http://www.ia.unc.edu/dev NeuroLib]
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:JointRegSeg.png|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:RegistrationRegularization|Optimal Atlas Regularization in Image Segmentation]] ==
| |
| − | | |
| − | We propose a unified framework for computing atlases from manually labeled data sets at various degrees of “sharpness” and the joint registration and segmentation of a new brain with these atlases. Using this framework, we investigate the tradeoff between warp regularization and image fidelity, i.e. the smoothness of the new subject warp and the sharpness of the atlas in a segmentation application.
| |
| − | [[Projects:RegistrationRegularization|More...]]
| |
| − | | |
| − | <font color="red">'''New:'''</font> B.T.T. Yeo, M.R. Sabuncu, R. Desikan, B. Fischl, P. Golland. Effects of Registration Regularization and Atlas Sharpness on Segmentation Accuracy. In Proceedings of MICCAI: International Conference on Medical Image Computing and Computer Assisted Intervention, 683-691, 2007. '''MICCAI Young Scientist Award.'''
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:ICluster_templates.gif|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:MultimodalAtlas|Multimodal Atlas]] ==
| |
| − | | |
| − | In this work, we propose and investigate an algorithm that jointly co-registers a collection of images while computing multiple templates. The algorithm, called '''iCluster''', is used to compute multiple atlases for a given population.
| |
| − | [[Projects:MultimodalAtlas|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> M.R. Sabuncu, M.E. Shenton, P. Golland. Joint Registration and Clustering of Images. In Proceedings of MICCAI 2007 Statistical Registration Workshop: Pair-wise and Group-wise Alignment and Atlas Formation, 47-54, 2007.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:GroupwiseSummary.PNG|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:GroupwiseRegistration|Groupwise Registration]] ==
| |
| − | | |
| − | We extend a previously demonstrated entropy based groupwise registration method to include a free-form deformation model based on B-splines. We provide an efficient implementation using stochastic gradient descents in a multi-resolution setting. We demonstrate the method in application to a set of 50 MRI brain scans and compare the results to a pairwise approach using segmentation labels to evaluate the quality of alignment.
| |
| − | [[Projects:GroupwiseRegistration|More...]]
| |
| − | | |
| − | <font color="red">'''New:'''</font> S.K. Balci, P. Golland, M.E. Shenton, W.M. Wells III. Free-Form B-spline Deformation Model for Groupwise Registration. In Proceedings of MICCAI 2007 Statistical Registration Workshop: Pair-wise and Group-wise Alignment and Atlas Formation, 23-30, 2007.
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Morphometric Measures and Shape Analysis ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" | [[Image:Basis membership.png|200px]]
| |
| − | | style="width:85%" |
| |
| − | | |
| − | == [[Projects:MultiscaleShapeAnalysis|Multiscale Shape Analysis]] ==
| |
| − | | |
| − | We present a novel method of statistical surface-based morphometry based on the use of non-parametric permutation tests and a spherical wavelet (SWC) shape representation. [[Projects:MultiscaleShapeAnalysis|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> D. Nain, M. Styner, M. Niethammer, J. J. Levitt, M E Shenton, G Gerig, A. Bobick, A. Tannenbaum. Statistical Shape Analysis of Brain Structures using Spherical Wavelets. Accepted in The Fourth IEEE International Symposium on Biomedical Imaging (ISBI ’07) that will be held April 12-15, 2007 in Metro Washington DC, USA.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:HippocampalShapeDifferences.gif|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:ShapeAnalysis|Population Analysis of Anatomical Variability]] ==
| |
| − | | |
| − | Our goal is to develop mathematical approaches to modeling anatomical variability within and across populations using tools like local shape descriptors of specific regions of interest and global constellation descriptors of multiple ROI's. [[Projects:ShapeAnalysis|More...]]
| |
| − | | |
| − | <font color="red">'''New:'''</font> Mert R Sabuncu and Polina Golland. Structural Constellations for Population Analysis of Anatomical Variability.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:FoldingSpeedDetection.png|200px|]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:ShapeAnalysisWithOvercompleteWavelets|Shape Analysis With Overcomplete Wavelets]] ==
| |
| − | | |
| − | In this work, we extend the Euclidean wavelets to the sphere. The resulting over-complete spherical wavelets are invariant to the rotation of the spherical image parameterization. We apply the over-complete spherical wavelet to cortical folding development [[Projects:ShapeAnalysisWithOvercompleteWavelets|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> B.T.T. Yeo, W. Ou, P. Golland. On the Construction of Invertible Filter Banks on the 2-Sphere. Yeo, Ou and Golland. Accepted to the IEEE Transactions on Image Processing.
| |
| − | | |
| − | P. Yu, B.T.T. Yeo, P.E. Grant, B. Fischl, P. Golland. Cortical Folding Development Study based on Over-Complete Spherical Wavelets. In Proceedings of MMBIA: IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis, 2007.
| |
| − |
| |
| − | |-
| |
| − | | |
| − | | |
| |
| − | | |
| |
| − | | |
| − | == [[Projects:ShapeBasedLevelSetSegmentation|Shape Based Level Segmentation]] ==
| |
| − | | |
| − | This class of algorithms explicitly manipulates the representation of the object boundary to fit the strong gradients in the image, indicative of the object outline. Bias in the boundary evolution towards the likely shapes improves the robustness of the segmentation results when the intensity information alone is insufficient for boundary detection. [[Projects:ShapeBasedLevelSetSegmentation|More...]]
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:Progress_Registration_Segmentation_Shape.jpg|200px]]
| |
| | | | | | | | |
| | | | |
| − | == [[Projects:ShapeBasedSegmentationAndRegistration|Shape Based Segmentation and Registration]] == | + | == [[ProjectWeek200706:CorticalThicknessForAutism|Cortical Thickness for Autism]] == |
| | | | |
| − | This type of algorithm assigns a tissue type to each voxel in the volume. Incorporating prior shape information biases the label assignment towards contiguous regions that are consistent with the shape model. [[Projects:ShapeBasedSegmentationAndRegistration|More...]]
| + | Our goal is to begin a longitudinal study of early brain development by cortical thickness in autistic children and controls (2 years with follow-up at 4 years). We want to be able to make statistical group comparisons. [[ProjectWeek200706:CorticalThicknessForAutism|More...]] |
| | | | |
| − | <font color="red">'''New: '''</font> K.M. Pohl, J. Fisher, S. Bouix, M. Shenton, R. W. McCarley, W.E.L. Grimson, R. Kikinis, and W.M. Wells. Using the Logarithm of Odds to Define a Vector Space on Probabilistic Atlases. Medical Image Analysis,11(6), pp. 465-477, 2007. <b>Best Paper Award MICCAI 2006 </b> | + | <font color="red">'''New: '''</font> Participation in the NA-MIC Summer 2007 Project Week. |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:Meanviews.png|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:ParticlesForShapesAndComplexes|Adaptive, Particle-Based Sampling for Shapes and Complexes]] ==
| |
| − | | |
| − | This research is a new method for constructing compact statistical point-based models of ensembles of similar shapes that does not rely on any specific surface parameterization. The method requires very little preprocessing or parameter tuning, and is applicable to a wider range of problems than existing methods, including nonmanifold surfaces and objects of arbitrary topology. [[Projects:ParticlesForShapesAndComplexes|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> J Cates, PT Fletcher, M Styner, M Shenton, R Whitaker, Shape modeling and analysis with entropy-based particle systems, IPMI 2007, pp. 333-345.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:UNCShape_OverviewAnalysis_MICCAI06.gif|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:ShapeAnalysisFrameworkUsingSPHARMPDM|Shape Analysis Framework using SPHARM-PDM]] ==
| |
| − | | |
| − | The UNC shape analysis is based on an analysis framework of objects with spherical topology, described mainly by sampled spherical harmonics SPHARM-PDM. The input of the shape analysis framework is a set of binary segmentations of a single brain structure, such as the hippocampus or caudate. These segmentations are converted into a shape description (SPHARM) with correspondence and analyzed via Hotelling T^2 two sample metric. [[Projects:ShapeAnalysisFrameworkUsingSPHARMPDM|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font>
| |
| − | | |
| − | * First version of Shape Analysis Toolset available as part of UNC Neurolib open source ([http://www.ia.unc.edu/dev/download/shapeAnalysis download]) , this is to be added to the NAMIC toolkit.
| |
| − | * Slicer 3 module for whole shape analysis pipeline in progress (based on BatchMake and distributed computing using Condor)
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:UNCShape_ShapeCorrespondence.png|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:PopulationBasedCorrespondence|Population Based Correspondence]] ==
| |
| − | | |
| − | We are developing methodology to automatically find dense point correspondences between a collection of polygonal genus 0 meshes. The advantage of this method is independence from indivisual templates, as well as enhanced modeling properties. The method is based on minimizing a cost function that describes the goodness of correspondence. Apart from a cost function derived from the description length of the model, we also employ a cost function working with arbitrary local features. We extended the original methods to use surface curvature measurements, which are independent to differences of object aligment. [[Projects:PopulationBasedCorrespondence|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font>
| |
| − | | |
| − | * Software available as part of UNC Neurolib open source ([http://www.ia.unc.edu/dev website])
| |
| − | * Evaluation on lateral ventricles, hippocampi, caudates, striatum, femural bone. Outperforms standard MDL on complex structures.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:overcomplete_vs_biorthogonal_wavelets.jpg|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:CorticalSurfaceShapeAnalysisUsingSphericalWavelets|Spherical Wavelets]] ==
| |
| − | Cortical Surface Shape Analysis Based on Spherical Wavelets. We introduce the use of over-complete spherical wavelets for shape analysis of 2D closed surfaces. Bi-orthogonal spherical wavelets have been proved to be powerful tools in the segmentation and shape analysis of 2D closed surfaces, but unfortunately they suffer from aliasing problems and are therefore not invariant to rotation of the underlying surface parameterization. In this paper, we demonstrate the theoretical advantage of over-complete wavelets over bi-orthogonal wavelets and illustrate their utility on both synthetic and real data. In particular, we show that the over-complete spherical wavelet transform enjoys significant advantages for the analysis of cortical folding development in a newborn dataset. [[Projects:CorticalSurfaceShapeAnalysisUsingSphericalWavelets|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 4, APRIL 2007
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:separating_loops.jpg|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:TopologyCorrectionNonSeparatingLoops|Topology Correction]] ==
| |
| − | | |
| − | Geometrically-Accurate Topology-Correction of Cortical Surfaces using Non-Separating Loops. We propose a technique to accurately correct the spherical topology of cortical surfaces. Specifically,we construct a mapping from the original surface onto the sphere to detect topological defects as minimal nonhomeomorphic regions. The topology of each defect is then corrected by opening and sealing the surface along a set of nonseparating loops that are selected in a Bayesian framework. The proposed method is a wholly self-contained topology correction algorithm, which determines geometrically accurate, topologically correct solutions based on the magnetic resonance imaging (MRI) intensity profile and the expected local curvature. Applied to real data, our method provides topological corrections similar to those made by a trained operator. [[Projects:TopologyCorrectionNonSeparatingLoops|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 4, APRIL 2007
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:UNCShape_CaudatePval_MICCAI06.png|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:LocalStatisticalAnalysisViaPermutationTests|Local Statistical Analysis via Permutation Tests]] ==
| |
| − | | |
| − | We have further developed a set of statistical testing methods that allow the analysis of local shape differences using the Hotelling T 2 two sample metric. Permutatioin tests are employed for the computation of statistical p-values, both raw and corrected for multiple comparisons. Resulting significance maps are easily visualized. Additional visualization of the group tests are provided via mean difference magnitude and vector maps, as well as maps of the group covariance information. Ongoing research focuses on incorporating covariates such as clinical scores into the testing scheme. [[Projects:LocalStatisticalAnalysisViaPermutationTests|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font>
| |
| − | | |
| − | * Available as part of Shape Analysis Toolset in UNC Neurolib open source ([http://www.ia.unc.edu/dev/download/shapeAnalysis download]).
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:qdec.jpg|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:QDEC|QDEC: An easy to use GUI for group morphometry studies]] ==
| |
| − | | |
| − | Qdec is a application included in the Freesurfer software package intended to aid researchers in performing inter-subject / group averaging and inference on the morphometry data (cortical surface and volume) produced by the Freesurfer processing stream. The functionality in Qdec is also available as a processing module within Slicer3, and XNAT. [[Projects:QDEC|More...]]
| |
| − | | |
| − | See: [http://surfer.nmr.mgh.harvard.edu/fswiki/Qdec Qdec user page]
| |
| | | | |
| | |} | | |} |
| − |
| |
| − | == fMRI Analysis ==
| |
| − |
| |
| − | === Functional Activation Analysis ===
| |
| − |
| |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" | [[Image:mit_fmri_clustering_parcellation2_xsub.png|200px]]
| |
| − | | style="width:85%" |
| |
| − |
| |
| − | == [[Projects:fMRIClustering|fMRI clustering]] ==
| |
| − |
| |
| − | In this project we study the application of model-based clustering algorithms in identification of functional connectivity in the brain. [[Projects:fMRIClustering|More...]]
| |
| − |
| |
| − | <font color="red">'''New: '''</font> P. Golland, Y. Golland, R. Malach. Detection of Spatial Activation Patterns As Unsupervised Segmentation of fMRI Data. In Proceedings of MICCAI: International Conference on Medical Image Computing and Computer Assisted Intervention, 110-118, 2007.
| |
| − |
| |
| − | |-
| |
| − |
| |
| − | | | [[Image:FMRIEvaluationchart.jpg|200px]]
| |
| − | | |
| |
| − |
| |
| − | == [[Projects:fMRIDetection|fMRI Detection and Analysis]] ==
| |
| − |
| |
| − | We are exploring algorithms for improved fMRI detection and interpretation by incorporting spatial priors and anatomical information to guide the detection. [[Projects:fMRIDetection|More...]]
| |
| − |
| |
| − | <font color="red">'''New:'''</font> Wanmei Ou, Sandy Wells, Polina Golland. Bridging Spatial Regularization And Anatomical Priors in fMRI Detection. In preparation for submission to IEEE TMI.
| |
| − |
| |
| − | |}
| |
| − |
| |
| − | === Algorithm and Software Infrastructure ===
| |
| − |
| |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" | [[Image:Brain-flat.PNG|200px]]
| |
| − | | style="width:85%" |
| |
| − |
| |
| − | == [[Projects:ConformalFlatteningRegistration|Conformal Flattening]] ==
| |
| − |
| |
| − | The goal of this project is for better visualizing and computation of neural activity from fMRI brain imagery. Also, with this technique, shapes can be mapped to shperes for shape analysis, registration or other purposes. Our technique is based on conformal mappings which map genus-zero surface: in fmri case cortical or other surfaces, onto a sphere in an angle preserving manner. [[Projects:ConformalFlatteningRegistration|More...]]
| |
| − |
| |
| − | <font color="red">'''New: '''</font> Y. Gao, J. Melonakos, and A. Tannenbaum. Conformal Flattening ITK Filter. ISC/NA-MIC Workshop on Open Science at MICCAI 2006.
| |
| − |
| |
| − | |}
| |
| − |
| |
| | == NA-MIC Kit == | | == NA-MIC Kit == |
| | | | |
| | === NAMIC Software Process === | | === NAMIC Software Process === |
| − | | + | # [[NA-MIC/Projects/NA-MIC_Kit/CMake_-_NAMIC_Kit_Building|CMake - NAMIC Kit Building]] (Kitware) |
| − | {| cellpadding="10"
| + | # [[NA-MIC/Projects/NA-MIC_Kit/CPack_-_NAMIC_Kit_Distribution|CPack - NAMIC Kit Distribution]] (Kitware) |
| − | | style="width:15%" |
| + | # [[NA-MIC/Projects/NA-MIC_Kit/Dart_2_and_CTest_-_Software_Quality|Dart 2 and CTest - Software Quality]] (GE, Kitware) |
| − | | style="width:85%" | | |
| − | | |
| − | |} | |
| | | | |
| | === Software Infrastructure === | | === Software Infrastructure === |
| − | | + | # [[NA-MIC/Projects/NA-MIC_Kit/Slicer3|Slicer 3]] (Isomics, GE, Kitware, UCSD, UCLA) |
| − | {| cellpadding="10"
| + | ## [[NA-MIC/Projects/NA-MIC_Kit/MRML|MRML]] (Isomics, Kitware, GE) |
| − | | style="width:15%" | | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Coordinate_Systems|Coordinate Systems]] (GE, Isomics) |
| − | | style="width:85%" | | + | ## [[NA-MIC/Projects/NA-MIC_Kit/IO_Unification|IO Unification]] (GE, Isomics) |
| − | | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Execution_Model|Execution Model]] (GE) |
| − | |} | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Grid_Computing|Grid Computing]] (UCSD, GE, Kitware, Isomics) |
| | + | # [[NA-MIC/Projects/NA-MIC_Kit/Licenses_Unification|Licenses Unification]] (Kitware, Isomics) |
| | + | # Toolkits |
| | + | ## [[NA-MIC/Projects/NA-MIC_Kit/KWWidgets|KWWidgets]] (Kitware) |
| | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Teem|Teem]] (Harvard/BWH) |
| | | | |
| | === Training & Dissemination === | | === Training & Dissemination === |
| − | | + | # [[NA-MIC/Projects/NA-MIC_Kit/Training_Material_and_Workshops_for_NA-MIC_Kit|Training Material and Workshops for NA-MIC Kit]] (MGH, BWH) |
| − | {| cellpadding="10"
| + | # [[NA-MIC/Projects/NA-MIC_Kit/Dissemination|Dissemination for NA-MIC]] (Isomics, Kitware, Harvard) |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |} | |
| | | | |
| | == Other Projects == | | == Other Projects == |
| | | | |
| − | {| cellpadding="10"
| + | # [[Non_Rigid_Registration|Non-Rigid Registration]] |
| − | | style="width:15%" | [[Image:vxl.gif|200px]]
| + | # [[NA-MIC/Projects/Diffusion_Image_Analysis/FreeSurfer_NRRD_IO|FreeSurfer NRRD IO]] |
| − | | style="width:85%" |
| + | # [[Algorithm:MGH:FreeSurferNumericalRecipiesReplacement|FreeSurfer Numerical Recipies Replacement]] |
| − | | + | # [https://www.slicer.org/wiki/Slicer3:Fluorescence_and_Electron_Microscopy_Support Slicer3 Fluorescence and Electron Microscopy Support] |
| − | == [[Projects:FreeSurferNumericalRecipiesReplacement|Numerical Recipies Replacement]] ==
| |
| − | | |
| − | Our objective is to replace algorithms using the proprietary Numerical Recipes for C source base in FreeSurfer in the efforts to open-source FreeSurfer. This project has been completed through the use of the open source packages VXL (VNL) and Cephes. This includes the complete replacement of all Numerical Recipes in C code, and the implementation of a battery of unit tests for each replaced function. Currently the open source release is at a beta stage, and 25 beta releases of the source have been made. We anticipate a complete open source release in first quarter 2008. [[Projects:FreeSurferNumericalRecipiesReplacement|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Completed
| |
| − | | |
| − | |}
| |